Two technologies together can help build and maintain genetic library for a global industry

Barcodes are short stretches of deoxyribonucleic acid (DNA) that act like identification strips of the produce we buy in supermarkets. These tiny stretches of genetic material, coming from almost any part of an organism, can be used to classify any number or kind of living things.
When two organisms appear to be similar on the surface, even small differences in their barcodes allow us to distinguish one from the other – a feat that was not possible over a century ago. People like Carolus Linnaeus, who undertook the challenge of classifying much of the natural world, had to look at the existing differences in organisms from their outward appearance, making judgements on what belonged where solely on physical traits. While not entirely wrong, many similar-looking organisms may be incredibly variable on a molecular scale, and barcoding allows us to see not just how they are different by using pre-existing information coming from what has been barcoded.
This means the more barcoded organisms we have, the easier it becomes to identify an unknown species, compare identities and delineate between groups of organisms. This becomes even more important when there is a vast difference in the morphological traits of an organism, including size, colors and other characteristics. For example, many fishes can have various developmental stages and visible phenotypic variation during their development, making recognition and classification burdensome and error-prone, leading to a variety of taxonomic and aquaculture problems.
The simplicity of DNA barcoding
The principle of DNA barcoding (DBC) is so simple and applicable that it has gained worldwide following for use for any species on the planet. The barcoding process is simple enough: Extract DNA, use it as a template to amplify a short sequence of the DNA called a marker in vitro through a technique called Polymerase Chain Reaction (PCR), and then uniquely sequence the PCR product from the selected organism.
Traditionally, there had been several markers used in DBC, coming from various DNA-containing organelles of cells such as the nucleus, mitochondria and chloroplasts (in the case of plants and other photosynthetic organisms). Then, in 2003, Professor Paul Hebert and his colleagues at the University of Guelph in Canada proposed the use of a single DNA marker, Mitochondrial cytochrome c oxidase subunit 1 (CO1), to be used for any species coming from any taxonomic group. The robustness of DBC was proven when it was seen to amplify even degraded DNA.

With estimates of more than 30,000 different fish species in the world today, with numerous shapes, characteristics and stages of growth, barcoding has become a routine tool in fish biodiversity and aquaculture research, prompting a campaign to DNA-barcode all fishes. Currently, this campaign finds its application through the fish barcode of life project (FISH-BOL) started in 2005, where roughly 25 percent of the 30,000 currently known fishes have been identified and at least one member of 89 percent of all known families has been barcoded.
One year after, the CO1 gene of roughly 8,000 fishes had been barcoded and more than 90 percent of fishes so far analyzed are apparently barcode-distinguishable. While the number is far from staggering, building a library on fish identity that is accessible online for the global community has huge impacts on the reach of easy-to-access information, even in remote areas where aquaculture is practiced.
Despite the apparent simplicity and ease of use of DBC along with the strides that have been accomplished due to its extensive use and development, there may be an even better technology: the rise of next generation nucleic acid sequencing technologies (NGS) that facilitate massive and parallel sequencing of nucleic acid information from almost all possible samples.
NGS can not only be used to sequence whole genomes – the entire collection of DNA data of an organism – but can also be used to tackle its transcriptome, the full range of messenger ribonucleic acid (mRNA) in a cell or in tissues, thus telling us which genes are turned on or off at any particular time, or after a particular treatment. With the massive mobilization of NGS in the last decade, combined with the sheer amount of raw data it can produce on an organism and the reduction in NGS costs, does DNA barcoding still occupy a niche in aquaculture research?
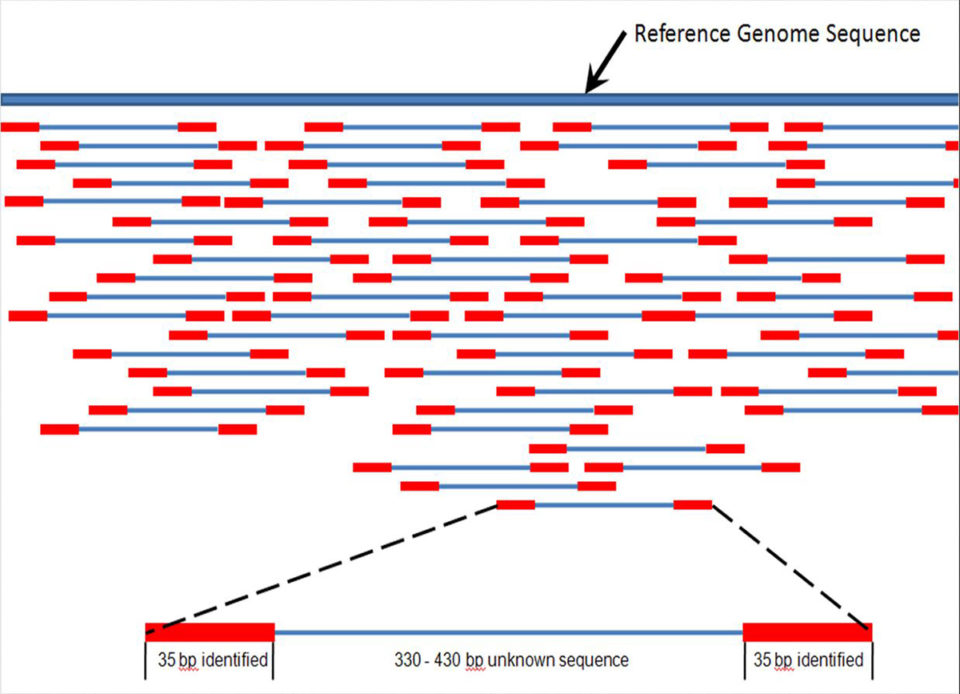
Barcoding and fish aquaculture
Barcoding in fishes has been traditionally used for fish biodiversity research. It has several strengths, such as in the identification of cryptic fish types and the determination of genetically indistinct populations, particularly in developing countries that are not as well equipped with the resources to perform more complicated methods of genetic screening and molecular identification. Despite this, DBC as a methodology still has much use in fish aquaculture and barcoding along with other DNA marker technologies have helped revolutionize aquaculture research.
Fishes are the largest vertebrate group, with more than 30,000 species, and provide a major role in feeding global economies and people. The proper identification of commercially important fish species is a basic and very necessary step in fisheries and aquaculture management, considering the number of variations of fish names given to similar species across different regions. With global aquaculture production increasing yearly and accounting for about half of global fish supplies, the proper identification to segregate commercially important fishes from morphologically indistinct counterparts is of vital importance for a variety of reasons:
- Perhaps the strongest use for DBC in aquaculture is identification that may prevent the mislabeling and substitution of fish products, especially those that undergo processing or are not so discernible, such as fish fillets. This has several potential impacts. First, mislabeled fish products can include fish with threatened biodiversity status and affect conservation efforts. Proper identification of market-sold fish may also prevent unwanted cases of allergies and health problems that arise from consumption of the wrong species due to confusing vernacular names, contributing both to health safety and food authentication. This becomes important in areas where there are several dialects that are often used to describe different species of fish. Species-specific contaminants in sold fish products – some of which may be harmful to human health, especially those found in raw fish – can also be detected in this manner.
- Hybrid identification is important for determining stock purity. This can influence farming practices and stringency of culture area, along with computing for and selecting against genetic drift (frequency change of an existing gene variant, or allele, in a population due to random sampling of its organisms). This is true not just for adult fish, but also for other life stages and their remains.
- Identification of wild broodstock through DBC is possible through the testing of eggs, larvae and identification of possible spawning areas where commercially important fish may reside. This is also true for estimating fishing quota in capture fisheries.
NGS in aquaculture
Despite fishes being the third most barcoded organisms globally and having a most relevant role in aquaculture, the use of DBC techniques in these animals has been affected by the progressive development of NGS technologies. NGS can process millions of small fragments of nucleic acid sequenced multiple times at a rate much faster than the traditional Sanger sequencing – the mainstay sequencing method for almost half a century – and covering a broader spectrum of information at increased sensitivity.
When considering the use of NGS over DBC, the amount of information that can be generated from using the technique is a very important factor. For example, one of the many platforms for NGS, the Roche 454 NGS platform, produces longer sequence reads than traditional DBC. The longer the reads are after sequencing, the better the picture created of the genome, with differences having the possibility of being highlighted better – an attainment that DBC has been striving to achieve for over a decade.
The next major factor is cost. While the simplicity and cost-effectiveness of DBC has always been one of its striking strengths, the cost of sequencing has become progressively lower, allowing more access to a variety of research groups and institutions. This greater coverage is also seen in the lower cost versions of equipment that are scaled down and are being made more affordable to more researchers.
The first NGS-derived full genome sequences for fish were made on model organisms, followed by commercially important fish; and the list continues to grow, now encompassing genomes from Atlantic Salmon, Antarctic cod, Japanese eel, Pacific bluefin tuna, catfish, common carp, tilapia, European sea bass, tongue sole, rainbow trout, pufferfish, Antarctic bullhead, Northern pike, Asian sea bass, grass carp, Asian arowana and spotted gar, among others.

These developments in computational technology are likewise being made available for use on a variety of aquatic organisms, including non-model and non-commercially important species. This opens the door for the use of NGS without the need for the reference data that DBC requires. Since DBC works on comparisons of existing data against the results of a new DBC analysis, then there is a bias where certain taxa benefit more than others. This may pose a problem for individuals exploring the use of non-barcoded individuals or those who have not yet received much attention as alternative sources of nutritive value for human and animal consumption.
Perhaps the strength of NGS compared to DCB is best illustrated by the flexibility of the use of its data that goes beyond identifying similarities and differences among taxa. NGS is fast becoming an important tool in finding genetic markers that may be related to genes of commercial interest to both suppliers and consumers, including genes that are of interest to innate immunity and fish physiology like antimicrobial peptides, probiotics and diet- and metabolism-interacting genes. These are necessary factors that contribute to the increase in aquaculture production and efficacy of its practices at various levels. Also, the whole genome pathogen sequencing of existing and emerging aquaculture pathogens and focusing on their epidemiology.
Perspectives: A combination of NGS and DBC?
Among aquaculture researchers, there is a great debate on the future of DBC, with some focusing on the more contemporary NGS approaches and labelling traditional genetic methods like DBC as outdated. Others take a more positive approach, citing the need for DBC to begin to incorporate the emergence of NGS into its methodologies.
The combination of DBC and NGS has been discussed, with the enhancement of DBC techniques being labelled as “metabarcoding” that can be used to detect environmentally discarded or degraded DNA (eDNA). eDNA has been used successfully in the estimation of distribution of invasive species in ponds, which may have a direct impact when dealing with invasive aliens that are piscivorous and decimate populations of commercially important cultured fish. eDNA use has also been investigated for applications in the marine environment using only seawater as a sample. While far from perfect, there have been efforts to detect extremely rare alleles, which may be useful for aquaculture. Rare alleles may be useful traits that can be increased through selective breeding and may be potential genetic resources to enhance fish production.
The union of DBC and NGS is still in its infancy and will need to be developed. Despite the arguments in favor of NGS, DBC still remains a staple aquaculture tool because of its capacity to be used in areas where the lowered cost of NGS is still a burden to the local population. Traditional DBC can still be used in countries that lack infrastructure and capital to devote resources into NGS in their research and fisheries management. While much time needs to be devoted in the reassessment of the use of barcoding in the current context of an NGS-driven genomics community, DBC can remain a staple tool for building the genetic library resources necessary for simple aquaculture needs such as species identification and differentiation.
References available from authors.
Authors
-
Zomesh A. Maini, M.Sc.
Department of Biology, School of Science and Engineering
Ateneo de Manila University
Katipunan Avenue, Loyola Heights
Quezon City, NCR, Philippines 1108 -
Vikas Kumar, Ph.D.
Division of Aquaculture, College of Agriculture, Food Science and Sustainable Systems
Kentucky State University
Frankfort, Kentucky 40601, USA
-
Janice A. Ragaza, Ph.D.
Corresponding author
Department of Biology, School of Science and Engineering
Ateneo de Manila University
Katipunan Avenue, Loyola Heights
Quezon City, NCR, Philippines 1108
Tagged With
Related Posts

Intelligence
Can a ‘chemical fingerprint’ deter seafood fraud?
Because paper trails aren’t perfect, some food producers are going beyond written or digital records to prove their products’ authenticity and prevent economic fraud. Is farmed seafood a perfect fit for forensic-science traceability?

Health & Welfare
Double-stranded RNA against WSSV genes provides antiviral protection in shrimp
Silencing genes in white spot syndrome virus (WSSV) with critical roles in replication could provide a strong antiviral effect and thus reduce shrimp mortality. The authors therefore established a study to evaluate the antiviral efficacy of double-stranded (ds)RNA against non-structural WSSV genes.

Responsibility
Genetic improvement aids red tilapia growth in Egypt
A new breeding program for genetic improvement of red tilapia was established at the Fish Research Center (FRC), Suez Canal University, in Ismailia, Egypt. It aims to improve the growth rate of the fish and to provide significant benefits to tilapia farmers.

Innovation & Investment
Genetic tool aims to stem the tide of oyster virus
Initially, a new chip loaded with oyster DNA pieces will identify oysters resistant to the herpes virus, but in the future it may be able to identify oysters with other desirable characteristics, such as faster growth rates.


