Results show in vitro antimicrobial potential against major fish pathogen

An alternative to the use of antibiotics in fish culture is the administration of probiotic bacteria, which have shown benefits in aquaculture both for water use sustainability and for fish health and productivity. Among other benefits, probiotic bacteria can improve the immune response and disease resistance of fishes and can promote growth. In recent years, due to the complexity of the intestinal microbiota of humans and other animals, probiotics composed of more than one bacterial strain have been developed.
The competitive exclusion culture is a technique by which a mixture of microorganisms from a healthy host are developed and maintained in order to selectively exclude enteropathogens (any organism causing intestinal tract disease). The competitive exclusion concept considers that the administration of intestinal microbiota from an adult animal to young animals could give protection against pathogenic bacteria.
Most competitive exclusion cultures have been applied to poultry, pigs and humans. In tilapia, a previous study showed that mixtures of different bacteria, in association, allowed upregulation (increase in cellular response to a molecular stimulus due to increased number of receptors on the cell surface) of immune responses and a reduction in mortality when challenged with the pathogenic bacteria Aeromonas hydrophila and Streptococcus iniae, as well as increased feed conversion ratio, although not improved the growth rate of tilapia. In another study, a commercial mixture of various bacteria, when applied continuously over six weeks, improved growth and enhanced tilapia intestinal immunological status.
This article – adapted and summarized from the original publication – reports the characterization of the microbial community composition and the establishment and dynamics of a continuous-flow competitive exclusion culture (CFCEC) derived from gut microbiomes of Nile tilapia (Oreochromis niloticus) specimens from tilapia farms in Colombia. To our knowledge, this is the first report of the development and characterization of a continuous-flow competitive exclusion culture derived from the tilapia gastrointestinal tract.
Study setup
Adult Nile tilapia were donated by commercial fish producers from two fish farms in different regions of Colombia. Fish were properly euthanized and transported to the laboratory, where their intestines were removed aseptically and samples collected and appropriately preserved, or used to develop a continuous-flow competitive exclusion culture (CFCEC) derived from gut microbiomes of Nile tilapia (Oreochromis niloticus) according to the method by Aguilar-Rivera et al. with some modifications.
The CFCEC was monitored over time to determine when the culture established a stable microbiota consortium. Parameters monitored were pH, total anaerobic and aerobic cell counts, bacterial diversity assessments, and antibacterial activity of CFCEC supernatant extracellular/released metabolites.
Samples were selected on days 0, 1, 2, 3, 5, 17 and 33 of the CFCEC to identify the bacterial composition variation by 16S rRNA gene amplicon Illumina sequencing [sequencing the bacterial 16S rRNA gene across multiple species is a widely used method for phylogeny and taxonomy studies].
For detailed information on the experimental design and continuous-flow competitive exclusion culture (CFCEC); characterization of the CFCEC; culture pH and total anaerobic and aerobic cell counts; characterization of the CFCEC bacterial diversity; analysis of the antibacterial activity of the CFCEC supernatant; and statistical analyses, refer to the original publication.
Results and discussion
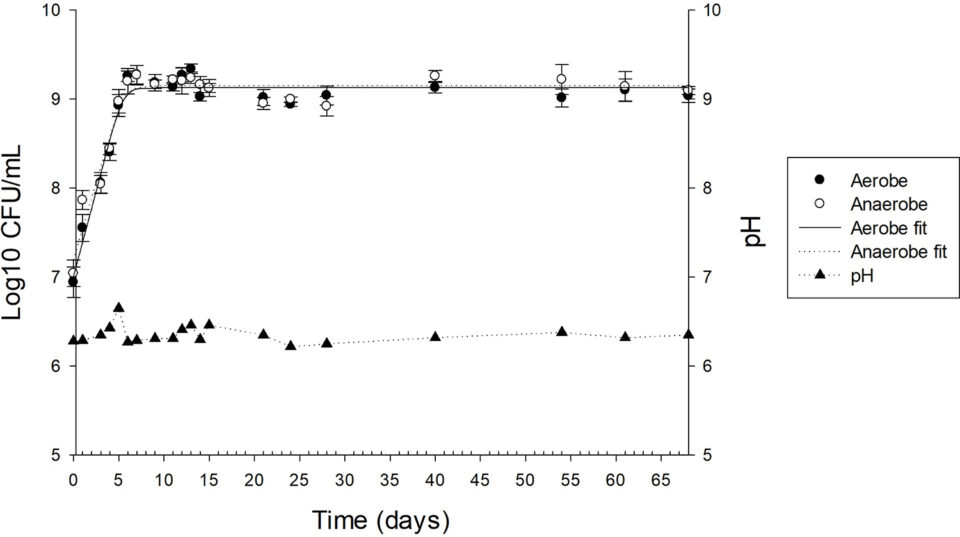
Results of our study allowed the identification of predominant bacterial genera in the continuous-flow competitive exclusion culture exhibiting antibacterial activity against the fish pathogen Streptococcus agalactiae. And data showed that aerobe and anaerobe growth parameters were similar, with an initial count (initial value) of around 107 CFU/mL (colony forming unites per milliliter, a unit used to estimate the number of viable bacteria or fungal cells in a sample) and a final count (final value) of more than 109 CFU/mL until day 68 and a maximum growth rate of less than 0.386/day (Fig. 1). The pH was stable, without external adjustments, between 6.25 to 6.35 during the 33 days evaluated.
Regarding the characterization of CFCEC bacteria, the bacterial taxonomic composition of the Nile tilapia intestinal contents showed that the intestinal contents from the two farms had differences in bacterial composition. In fish from the farm in the Atlantic region, the most abundant phylum was Firmicutes and the genus Clostridium. In contrast, for fish from the Llanos (inland plains) region, the most abundant genus was Cetobacterium from the phylum Fusobacteria followed by Clostridium and Plesiomonas (Fig. 2).
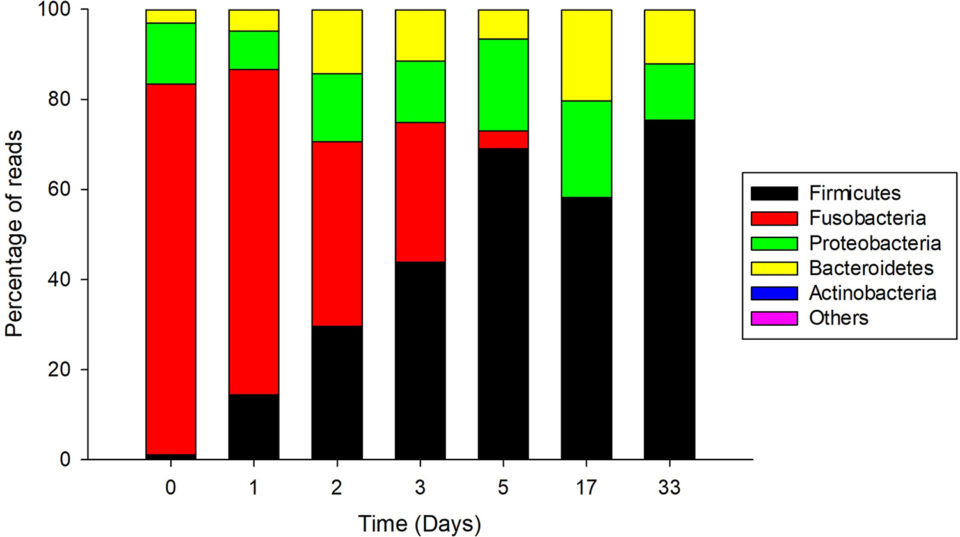
In the CFCEC, we observed an increase in the relative abundance of lactic acid bacterial types. Lactococcus was the most abundant genus at 33 days of the CFCEC. However, despite the well documented antibacterial effects of this genus, medium from the 33-day CFCEC did not exhibit agar diffusion anti-S. agalactiae activity. Lactococcus is generally recognized as safe (GRAS) and therefore commonly used in food industry. The probiotic potential of this genus to various cultured fish – including various groupers, cobia, tilapia, carps and others – has been reported, including growth promotion, antibacterial activity and immunomodulation. Lactococcus has also been applied to cultured salmon and turbot as a probiotic because of its immunomodulatory properties.
There are different antimicrobial mechanisms ascribed to competitive exclusion: biological (low levels of oxygen), chemical (organic acids), biochemical (antimicrobial substances) and nutritional (competition for nutrients). In the first three days of our experiment, the cell-free extracellular products of the CFCEC exhibited an antibacterial activity against S. agalactiae. Given this result, we consider it is very likely that the CFCEC harbored bacterial types able to produce antibacterial substances against this fish pathogen.
However, considering the loss of antibacterial activity from day three in the culture supernatant, it is important to evaluate a competitive exclusion and displacement of pathogens by the CFCEC consortium in a chemostat or by in vivo challenging of fishes in order to identify whether other mechanisms are involved in the inhibition of pathogenic bacteria by the CFCEC derived from tilapia gut microbiomes.
Perspectives
Results of our study showed that the intestinal microbiota of Nile tilapia in the CFCEC demonstrated in vitro antimicrobial potential against the fish pathogen S. agalactiae. And the 16S rRNA gene amplicon sequencing revealed the abundant bacterial types at different stages of the CFCEC establishment, with compositional changes observed over time that were related to the increase or decrease of anti-Streptococcus agalactiae activity.
Additional characterization of the CFCEC is needed to evaluate the competitive exclusion and displacement potential against S. agalactiae and to identify the mechanisms used by the probiotic bacteria in order to inhibit the growth of pathogenic bacteria. For further in vivo studies, it will be necessary to develop a CFCEC culture selecting bacteria based on probiotic properties and metabolic compatibility.
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-
Javier Fernando Melo-Bolívar, Ph.D.
Universidad de La Sabana, Facultad de Ingeniería
Campus Universitario del Puente del Común
Km 7 Autopista Norte de Bogotá, Chía, Cundinamarca, Colombia -
Ruth Yolanda Ruiz Pardo, Ph.D.
Universidad de La Sabana, Facultad de Ingeniería
Campus Universitario del Puente del Común
Km 7 Autopista Norte de Bogotá, Chía, Cundinamarca, Colombia -
Michael E. Hume, Ph.D.
United States Department of Agriculture
Agricultural Research Service
Southern Plains Agricultural Research Center
College Station, TX USA -
David J. Nisbet, Ph.D.
United States Department of Agriculture
Agricultural Research Service
Southern Plains Agricultural Research Center
College Station, TX USA -
Fernando Rodríguez-Villamizar, Ph.D.
Corporación Colombiana de Investigación Agropecuaria (Agrosavia)
Centro de Investigación Tibaitatá, Mosquera, Cundinamarca, Colombia -
Juan F. Alzate, Ph.D.
Centro Nacional de Secuenciación Genómica-CNSG
Sede de Investigación Universitaria SIU
Grupo de Parasitología, Facultad de Medicina
Universidad de Antioquia, Medellín, Colombia -
Howard Junca, Ph.D.
Microbiomas Foundation, Div. Ecogenomics & Holobionts
RG Microbial Ecology: Metabolism, Genomics & Evolution
Chía, Colombia -
Luisa Marcela Villamil Díaz, Ph.D.
Corresponding author
Universidad de La Sabana, Facultad de Ingeniería
Campus Universitario del Puente del Común
Km 7 Autopista Norte de Bogotá, Chía, Cundinamarca, Colombia
Tagged With
Related Posts

Health & Welfare
Antimicrobial resistance: Aquaculture to humans, humans to aquaculture?
The transfer of resistance from an aquaculture pathogen into a human enteric bacterium has been demonstrated in research and may be happening in the real-world environment.

Health & Welfare
Challenging Pacific white postlarvae with AHPND
Study results indicate that P. vannamei challenged with AHPND in biofloc had higher survival rates than shrimp challenged in clear water.

Health & Welfare
Salmon scientists collaborate on new global health initiative QASH
A cooperative effort between salmon scientists in Norway, Scotland, Chile and Canada aims to develop a globally applicable tool to help management and certification of salmon farms.

Health & Welfare
Establishing nitrifying bacteria in super-intensive biofloc shrimp production
In biofloc production systems, ammonia-oxidizing bacteria process ammonia to nitrite, which provides substrate for nitrite-oxidizing bacteria that process nitrite to relatively non-toxic nitrate.


