Implementing practical measures at the hatchery to reduce family skews

Effective breeding programs seek to maintain genetic diversity to avoid issues from inbreeding and allow long-term selection of desirable production traits. In contrast to terrestrial agriculture species, many aquaculture species are highly fecund, mass spawned, and communally reared, which results in skewed [asymmetric] family contributions which pose challenges to breeding programs.
Skewed family distributions are common in aquaculture species that are highly fecund, communally (mass) spawned, and/or communally reared. The magnitude of skews pose challenges for maintaining family-specific genetic diversity, as increased resources are required to detect individuals from underrepresented families, or reliably determine relative survival as a measure of family performance.
In most aquaculture species, it is impractical to track families identified from independent spawnings through physical tagging methods, due to their small body size and high fecundity. While it is possible to spawn and rear families individually at the hatchery, this requires significant resources and introduces additional environmentally induced rearing effects among families.
An effective alternative to trace and identify families is the use of genetic markers for parentage assignment. These genetic approaches have proven reliable to recover important genealogical information from communally reared families. Genetic markers have been used for parental assignment and family distributions in a number of important aquacultured species that have been communally reared.
There is limited understanding of family skews or changes in family proportion of communally reared shrimp under commercial rearing conditions and particularly how this may affect genotyping strategies to recover family performance data in breeding programs. The black tiger shrimp, P. monodon, is an example of a highly fecund aquaculture species, which commonly displays order of magnitude differences in the production of viable progeny from each spawn.
This article – adapted and summarized from the original publication (Foote A., D. Simma, M. Khatkar, H. Raadsma, J. Guppy, G. Coman , E. Giardina, D. Jerry, K. Zenger and N. Wade. 2019. Considerations for Maintaining Family Diversity in Commercially Mass-Spawned Penaeid Shrimp: A Case Study on Penaeus monodon. Front. Genet. 10:1127.) – reports on a study that determined family skews in P. monodon from three batches (separate spawning events) at two ages: 30 days of culture (DOC) prior to stocking into commercial grow-out ponds as well as 150 DOC at harvest age.
https://www.aquaculturealliance.org/advocate/unleashing-the-genetic-potential-of-black-tiger-shrimp/
Study setup
Wild P. monodon broodstock were sourced off the coast of Northern Territory, Australia, and transferred to a commercial hatchery at Flying Fish Point, Queensland. Broodstock underwent routine commercial maturation: maintained in an indoor tank system at a density of three per square meter, with flow through seawater maintained at 28±0.5 degrees-C and fed on a commercial maturation diet.
A total of 678 potential broodstock parents were allowed to mate naturally within the tank, with any unmated females then artificially inseminated. Females were unilaterally eyestalk ablated and those with ripe ovaries placed into communal spawning tanks, with up to five other mated females at any given time. As spawners were pooled, there was no equalization of family contribution; there was also no equalization across spawning tanks. Broodstock were not tagged, so the contributing parents were not known at the time of spawning; instead, genotyping would determine parents retrospectively.
Spawned eggs underwent routine commercial washing prior to transfer into a hatching tank, and then hatched nauplii were harvested and transferred into larval rearing tanks (LRTs) where they would be reared on a commercial diet to 30 DOC. At 30 DOC, LRTs were pooled, and progeny were stocked into commercial grow-out ponds, under routine commercial conditions at a density of approximately 45 per square meter until harvested at 150 DOC. The three separate batches of families were kept discrete, and the three batches of contributing parents, larval tanks, and grow-out ponds were tracked and sampled for genotyping and parental assignments.
The three batches of mass-spawned P. monodon were sampled at the two life stages (30 and 150 DOC), along with the broodstock, to determine family origin. All tissues for genetic analyses were fixed and the total nucleic acid (TNA) was extracted. The genetic material from each individual broodstock, as well as each individual progeny at both 30 and 150 DOC, was associated with a unique plate location and identification label, enabling tracking of individual tissue throughout the extraction, genotyping and parental assignment process.
For detailed information on animal origin, spawning and rearing; genetic sampling; nucleic acid extraction and genotyping; family assignment; relative family contribution; and sampling effort under skewed family contributions, refer to the original publication.
Results and discussion
Overall, a total of 199 out of 678 potential parents contributed to the offspring sampled. A total of 2,914 individuals were sampled at 30 DOC and 2,820 at 150 DOC, with an overall assignment rate [statistical technique to determine the relationship between individuals and populations, to assign reference populations as origins of individuals] of 98.6 percent at 30 DOC and 99.3 percent at 150 DOC. The minimum assignment rate was 97 percent (LRT c), while the average assignment rate for each batch was 99.5 percent, 98.1 percent and 98.6 percent in batches 1, 2 and 3, respectively.
Families that had a contribution significantly different from zero were designated “top tier” for each batch, while the remainder of families had too few individuals to be reliably detected or further statistical analysis performed. Overall, the number of top tier families were 33 of 54 families in batch 1, 22 of 35 families in batch 2 and 24 of 33 families in batch 3.
The sample size of approximately 500 individuals per pond, 1,000 per batch, at two commercially important time points for P. monodon, was adequate to detect and redetect all but the rarest of families. This enabled the skewness of family distributions to be determined, changes in relative contribution of families and differences in family rankings at harvest between ponds stocked with the same families. However, breeding programs focused on genetic improvement would typically benefit from increased sample sizes to improve the number of families detected and the accuracy of performance evaluations for each family. The benefits of increased sample sizes and sampling strategies as well as practical hatchery practices are discussed further below.
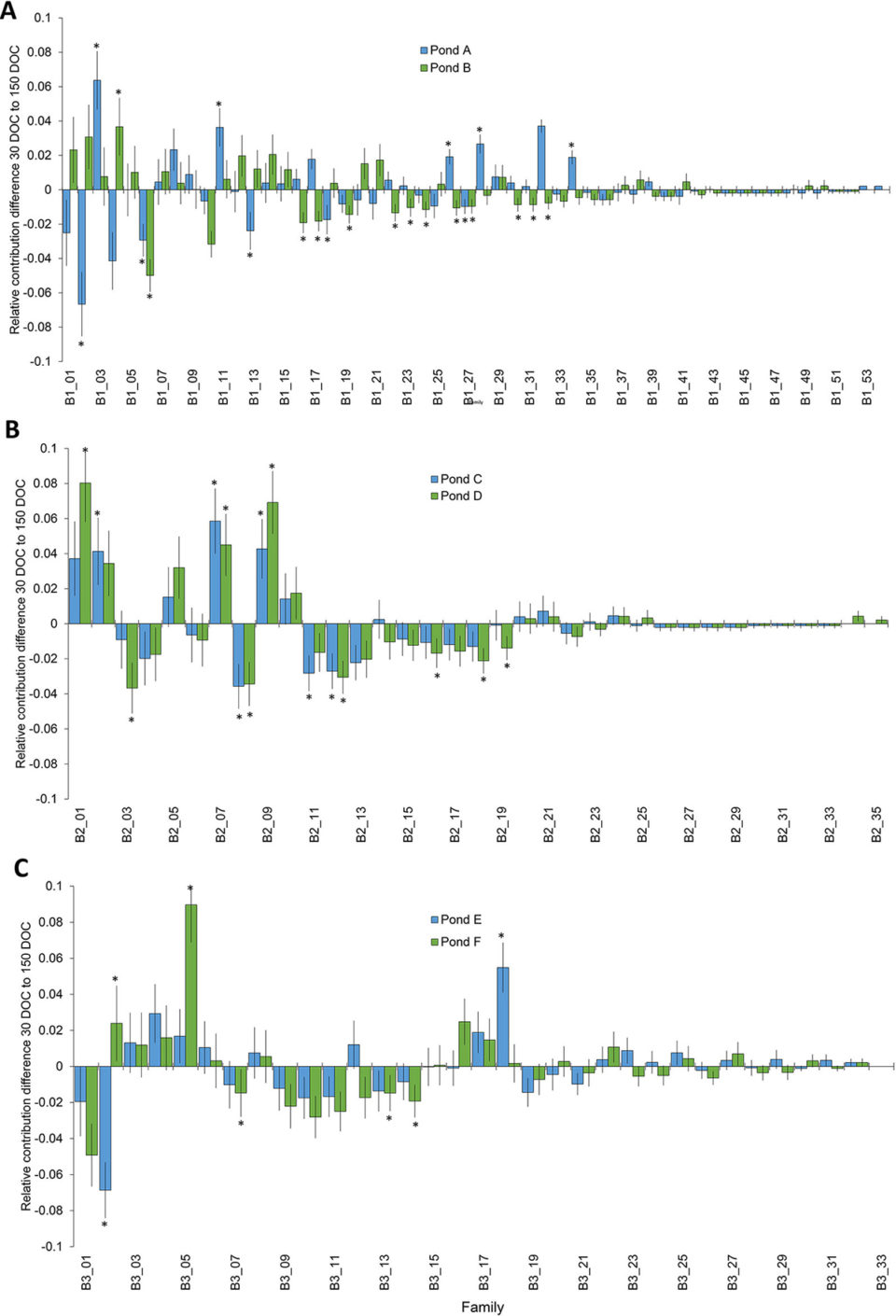
Sampling approximately 500 individuals per pond was sufficient to recapture all of the top tier represented families detected at pond stocking (30 DOC), again at harvest (150 DOC), in four of six ponds from two of three batches. When the two ponds from the remaining batch were combined, the batch sample size of approximately 1,000 was sufficient to capture all families in the top tier at both time points. The overall skewness of mass-spawned P. monodon in the current study increased from pond stocking (30 DOC), where relative family contributions were <1 to 11 percent, to harvest (150 DOC), where relative family contributions were <1 to 18 percent. The magnitude of family skews in the current study was similar to other aquaculture species including other penaeid shrimp.
Results demonstrated that differences in family skews, incorporating the effects specific to each species and the sampling time points in the production cycle, should be considered when determining sampling and genotyping effort to reliably determine family diversity within breeding programs.
To understand the relative impact of family skews and develop mitigation strategies, the underlying causes of skews needs to be determined. The skews at stocking for each batch (maximum 17.7 percent relative family contribution) was higher than the difference in skews from stocking to harvest for each batch (maximum 9 percent relative family contribution), indicating that initial factors such as variable broodstock fecundity and larval survival had more of an impact on family skews than differential survival during pond grow-out.
Previous studies have reported that hatchery practices and differential family survival in that hatchery production stages can result in dramatic reductions in the effective population size (by greater than 70 percent) within a single generation. Practical strategies could be employed by hatcheries to lessen family skewing up to pond stocking, such as individually spawning and equal pooling of progeny per family at various time points in the hatchery. While pooling of families closer to the point of pond stocking will result in more equal family distributions, keeping families separate to trace family origin also has the potential to introduce a range of environmental rearing effects.
Depending on the breeding program requirements, the best and most practical compromise might be to mitigate the variable fecundity, by spawning individually and equalizing contributions by pooling families once the progeny have hatched.
The change in relative family proportions from pond stocking to harvest can be used as a measure of relative family survival, allowing ranking of families and a potential selection criterium in breeding programs. While changes in relative family proportions from stocking to harvest were observed for many families, the sample size of approximately 500 per pond only resulted in detection of significant changes in proportion for 31 percent of the top tier and 20 percent of the total families.
If this relative survival is to be used as a performance measure, the sample size would need to increase to allow detection of significant changes in a greater proportion of families. Increasing the sample size to 5,000, for example, would allow detection of significant changes in 78 percent of families. Cost-effective strategies to increase the quantity of individuals genotyped might be facilitated in the future through strategies such as DNA pooling that has been validated in other production animals such as chicken, beef cattle and sheep. DNA pooling allows significant cost savings with a number of pooled individuals genotyped together per sample.
There was little consistency between the relative survival of families, as well as the ranking of families by relative contribution across ponds, for each of the three batches. However, the majority of families in the top and bottom tiers of abundant families was consistent across ponds. This difference in family survival performance across ponds highlights the impact of environmental rearing effects, in addition to genetic effects. A future trial could stock families evenly across replicate ponds to discriminate the magnitude of genotype by environment (G×E) interactions influencing relative family survival) and their changes based on the genetic merit of the families.
Inbreeding would also be more easily controlled in mass spawning approaches where broodstock are tagged and their pedigree/relatedness known, as this would allow informed mating crosses and tracking of contributions to progeny. However, it must be acknowledged that in a communal spawning and rearing approaches, it is more difficult to track smaller families, and they are more likely to be lost.
Perspectives
Overall, skewed relative family contributions increased from pond stocking (30 DOC) to harvest age (150 DOC). However, skewness at pond stocking was greater than the increase in skewness during pond grow-out. Therefore, practical mitigation strategies implemented in the hatchery are likely to reduce overall skewing; however, the cost, practicality, and differential environmental rearing effects of the strategy implemented need to be considered. Equalizing and pooling family contributions after they have hatched as nauplii may be a cost-effective strategy.
The change in relative family proportion from stocking to harvest age provided a survival measure and family ranking to be determined for each family, which could be incorporated as a selection criterium in breeding programs. Differences in survival and family rankings between ponds outline the importance of environmental effects on survival. The sample size of approximately 500 individuals genotyped per pond was sufficient, with the skewness observed, to detect most families at harvest age.
However, larger sample sizes would allow greater numbers of individuals per family to be detected as well as increase the ability to detect families that showed a significant change in relative proportion. Models that incorporated the family skews demonstrated the magnitude increase in sampling required to detect the lowest 25 percent of families as well as their change in proportion during pond grow-out.
Overall, this study provides the practical information on sampling effort to detect families accurately and reliably for future selective breeding programs in highly fecund aquacultured species including shrimp and recommendations on key actions to mitigate skews.
References available in the original publication.
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Author
-
Andrew Foote, Ph.D.
Corresponding author
ARC Research Hub for Advanced Prawn Breeding, James Cook University, Townsville, QLD, Australia; and
Centre for Sustainable Tropical Fisheries and Aquaculture, College of Science and Engineering, James Cook University, Townsville, QLD, Australia; and
Aquaculture Program, CSIRO Agriculture and Food, Queensland Bioscience Precinct, St. Lucia, QLD, Australia
Editor’s note: This article has 10 co-authors, but only the corresponding author is listed.
Related Posts

Health & Welfare
Domestication of Pacific white shrimp revolutionizes aquaculture
The development of specific pathogen-free Pacific white shrimp and breeding led to rapid adoption of the domesticated shrimp throughout the Western Hemisphere and Asia.

Aquafeeds
Global brine shrimp supply a potential bottleneck to aquaculture expansion, part 1
The brine shrimp artemia is critical for larval fish and shrimp diets. As aquaculture expands, an effective artificial replacement diet for artemia is a major consideration for the industry.

Health & Welfare
Broodstock shrimp nutrition, a review
Research into broodstock shrimp nutrition will be a key element in the further use of domesticated and genetically selected stocks for aquaculture.

Intelligence
Obstacles and opportunities in penaeid shrimp breeding
As economic benefits of selective penaeid shrimp breeding become more compelling, the shrimp industry will invest in breeding programs to produce genetically superior stocks.


