Correct parents could be identified with 98 and 92 percent accuracy

Shrimp, like most aquaculture species, are great candidates for achieving rapid gains through selective breeding. Their high fecundity generates a broad gene pool from which elite performers can be selected.
One of the key problems in shrimp-breeding programs is how to track pedigree and assess the relative performance of individual families and family groups in communal environments without physically tagging the animals. Molecular markers allow producers to maximize the gains from selective breeding while monitoring inbreeding.
The recent domestication of the black tiger shrimp, (Penaeus monodon), will allow the development of selective-breeding programs for the species. DNA marker technologies can then be utilized to carry out pedigree tracking and family assessments that ensure the selective breeding is effective.
DNA fingerprinting
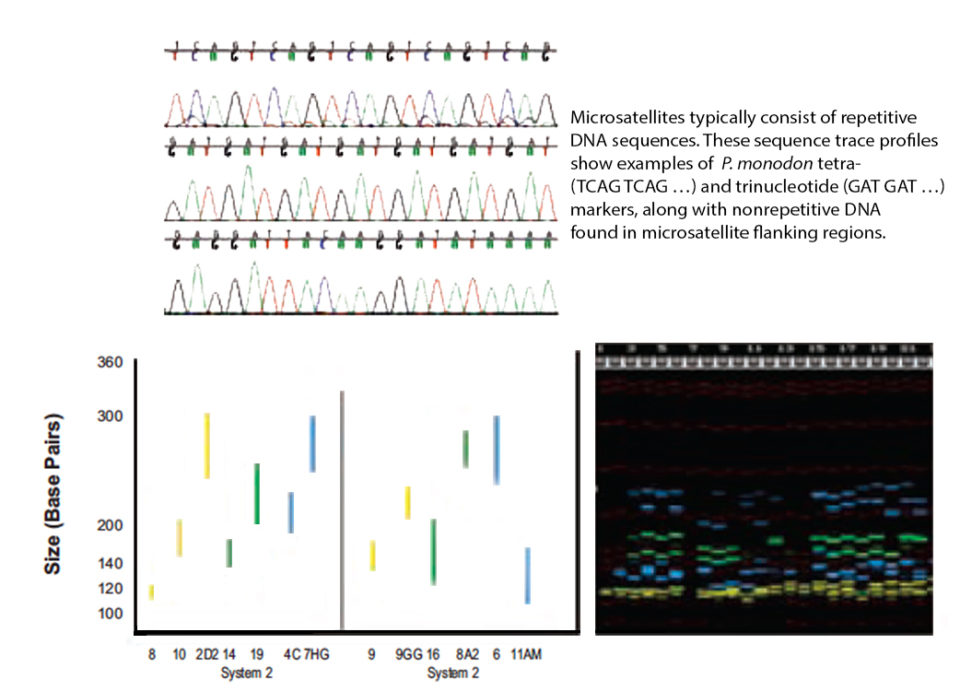
DNA fingerprinting using micro-satellites has wide-ranging applications in the field of genetics, including investigations of genetic variability, inbreeding, parentage assignments, species and strain identification, and the construction of high-resolution genetic linkage maps. They typically consist of repetitive DNA sequences of two, three or four nucleotides that can be repeated six to 100 times.
Microsatellites usually have no direct effect on production traits and have been widely used in breeding programs for numerous livestock and aquaculture species. Once the markers are sufficiently characterized to determine allele numbers and size ranges, they can also be multiplexed to create a fingerprinting “panel,” whereby a number of markers (usually five to 10) are each labeled with a different fluorescent dye but scored simultaneously. Multiplex systems therefore result in significant savings both in time and money.
Design considerations
With the aim of designing a robust Penaeus monodon microsatellite marker system for use in DNA fingerprinting in the Australian prawn aquaculture industry, the authors chose to impose a number of strict criteria for the types of microsatellite markers used. The markers had to be easy to score, with consistent amplification across genetically diverse populations, and be suitable for multiplex systems.
With access to wild animals from Australian waters and farmed animals from Thailand, the authors assessed the criteria above to generate reproducible, low-cost and high-throughput systems for on-farm applications such as assessment of broodstock diversity and tracing parentage of high-performing families.
Assessing preexisting markers
Prior to the study, there were already 184 putative microsatellites for P. monodon in public DNA databases. These microsatellite markers had primarily been developed for studying genetic population structures and not been used in multiplex systems. Over two-thirds of the microsatellites contained dinucleotide repeats.
To meet the first criterion of ease of scoring, the authors chose to ignore all dinucleotide microsatellites, concentrating rather on tri- and tetranucleotide repeats. Microsatellite markers with dinucleotide repeats are notorious for exhibiting “stutter” bands that make scoring complicated and less accurate. Other considerations for choosing markers were the suitability of flanking sequence for primer design and expected product sizes in the range of 100-350 bp.
Of the 184 publicly available markers, only 19 were deemed suitable for further assessment. The authors tested the markers across a small panel of 15 shrimp, 10 from Australian waters and five from Thailand. Seven markers did not amplify the Australian shrimp, reflecting their origins (i.e., developed using Asian populations). Twelve were retained for further evaluation.
New microsatellites
With only 12 of the public micro-satellite markers worthy of further investigation, the authors decided to isolate new markers. Of 42 unique tri- and tetranucleotide microsatellite sequences obtained, eight sequences were assessed on the small panel of animals. Seven markers looked promising and were retained for further evaluation.
Designing multiplex systems
The 19 candidate markers were next individually assessed on a larger panel of 93 Australian and 64 Thai shrimp to investigate size range and number of alleles, their ease of amplification and reliability of allele calling. The multiplex technology available at the time of the study meant there were three fluorescent dyes available for labeling the markers.
Dyes can be used more than once so long as no two markers labeled with the same dye have overlapping size ranges. Based upon size range and compatibility of the fluorescent dyes, 13 markers were combined into two multiplex systems with six and seven markers, respectively, as shown schematically in Fig. 1.
Ongoing studies
With the systems now developed, the authors began to apply them in earnest for on-farm applications. The ability to identify parents and assign pond-reared offspring to appropriate families and family groups is a major focus of the current work.
A “blind” tank trial using 120 animals from known families demonstrated that the correct mothers and fathers could be identified with 98 and 92 percent accuracy. This was a very promising result that opens the way for on-farm selection of high-performance families without the need to use costly alternatives such as separate rearing of families/groups of families or tagging animals.
Other activities include the evaluation and monitoring of the genetic diversity of domesticated black tiger shrimp in Australia relative to wild populations, assessment of family structure and dynamics within commercial ponds, and comparison of genetic diversity within and across different Australian and international populations of P. monodon.
(Editor’s Note: This article was originally published in the March/April 2008 print edition of the Global Aquaculture Advocate.)
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-
Russell Lyons, Ph.D.
CSIRO Food Futures Flagship
CSIRO Livestock Industries – Level 5, QBP
306 Carmody Road
Saint Lucia, Queensland 4067 Australia -
Leanne Dierens
CSIRO Food Futures Flagship
CSIRO Livestock Industries – Level 5, QBP
306 Carmody Road
Saint Lucia, Queensland 4067 Australia -
Tom Dixon, Ph.D.
CSIRO Food Futures Flagship
CSIRO Livestock Industries – Level 5, QBP
306 Carmody Road
Saint Lucia, Queensland 4067 Australia -
Yutao Li, Ph.D.
CSIRO Food Futures Flagship
CSIRO Livestock Industries – Level 5, QBP
306 Carmody Road
Saint Lucia, Queensland 4067 Australia -
Nigel Preston, Ph.D.
CSIRO Food Futures Flagship
CSIRO Marine and Atmosphere Research
Cleveland, Queensland, Australia
Tagged With
Related Posts

Intelligence
A brief look at genetically modified salmon
If approved by FDA, fast-growing genetically modified salmon will provide a safe and nutritious product similar to other farmed Atlantic salmon.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 2
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors considered biotic and abiotic factors. Part 2 describes results of their study.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 3
In this third and final part, authors present recommendations to help reduce the incidence of Zoea-2 Syndrome, which is not caused by any known infectious agents in P. vannamei hatcheries in India.


