DNA sequences reacted with OIE-listed IHHN virus diagnostic primers
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) is one of the eight reportable crustacean pathogens listed in the World Organization for Animal Health (OIE) Aquatic Code. In some countries, exclusion and regulatory action are applied if polymerase chain reaction (PCR) tests are positive for IHHNV in samples of farmed or imported live shrimp.
In a 2006 article published in Virus Research, Drs. Kathy Tang and Donald Lightner reported on the occurrence of IHHNV-related DNA sequences within the genome of Penaeus monodon shrimp. The shrimp had been collected from locations in Australia, the Western Indo-Pacific and off the east coast of Africa.
In the 2012 online edition of the OIE Aquatic Manual of Diagnostic Tests for Aquatic Animals, the authors stated there are three distinct genotypes recognized for IHHNV. Types 1 and 2 are infectious viruses, while Type 3 is non-infectious and recognizable by conventional PCR testing, which detects the IHHN-related DNA sequences present in the genome of P. monodon.
The purpose of this article is to provide additional documentation on the geographic range for P. monodon where IHHN-related genomic DNA sequences can be expected in PCR tests using recommended primers, as well as provide additional information that demonstrates the sequences are inherited by offspring from their parents according to Mendel’s Law of Segregation.
Diverse distribution
Moana Technologies, LLC’s closed herd of Penaeus monodon – housed in the Moana Nucleus Breeding Center (NBC) in Kona, Hawaii, USA – is made up of offspring several generations removed from the original wild, adult founder parents sourced from the South China Sea, Andaman Sea, Bay of Bengal and off the east coast of Africa in the Indian Ocean. IHHN-related genomic sequences are a common feature in Moana monodon shrimp.
As an example of a “typical” distribution pattern for IHHN-related genomic sequences in Moana monodon, a listing of PCR test results using a series of IHHNV primers is presented in Table 1. The data presented are for a random sample of 20 sub-adult Moana monodon from a grow-out tank in the Moana NBC, a preserved sample of IHHNV-infected postlarval Litopenaeus vannamei from a hatchery in Asia, and an IHHNV-positive control plasmid courtesy of the Aquaculture Pathology Group at the University of Arizona, USA.
Brock, Summary of PCR test results for five IHHN primers, Table 1
| Sample | Generation | Sex | SMP-DNA | SYBR/ABP Bactim | IHHNV (309F/R) | IHHNV (389F/R) | IHHNV (392F/R) | IHHNV (77012/ 77353) | IHHNV SYBR/ABP |
|---|
Sample | Generation | Sex | SMP-DNA | SYBR/ABP Bactim | IHHNV (309F/R) | IHHNV (389F/R) | IHHNV (392F/R) | IHHNV (77012/ 77353) | IHHNV SYBR/ABP |
|---|---|---|---|---|---|---|---|---|---|
| A1 | f4 | F | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A2 | f6 | M | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A3 | f4 | F | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A4 | f6 | F | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A5 | f4 | M | Pos. | Pos. | Neg. | Pos. | Pos. | Neg. | Neg. |
| A6 | f5 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A7 | f6 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A8 | f5 | F | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A9 | f5 | F | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A10 | f6 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A11 | f6 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A12 | f6 | M | Pos. | Pos. | Neg. | Pos. | Pos. | Neg. | Neg. |
| A13 | f6 | F | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A14 | f6 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| A15 | f6 | M | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A16 | f6 | F | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A17 | f5 | M | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A18 | f6 | F | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A19 | f5 | M | Pos. | Pos. | Neg. | Neg. | Pos. | Neg. | Neg. |
| A20 | f6 | M | Pos. | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. |
| B1 | L.v. | N.A. | Pos. | Pos. | Neg. | Pos. | Pos. | Pos. | Pos. |
| IHHNV-positive control (plasmid) | IHHNV-positive control (plasmid) | N.A. | N.A. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
The data from 2009 show the PCR reaction status for seven conventional primers. Included were two quality-control primers, SMP-DNA shrimp muscle primer and an in-house DNA primer set, and the SYBR IHHNV kit Bactim primers. Four of the five IHHNV primers are listed in the OIE Aquatic Manual as suitable for detection of IHHNV. The fifth was a commercial IHHNV kit primer, SYBR/ABP IHHNV kit.
Note that for the Moana monodon samples, 50 percent of the shrimp were reactors to the 392F/R primers, 10 percent were PCR positive with the 389F/R primers, and none were PCR positive for the 309F/R, 77012/77353 or SYBR/ABP kit primers for IHHNV. Typically, the authors have found that individual shrimp that are PCR positive with the 389F/R primers are also positive with the 392F/R primers. All of the 20 Moana monodon tissue samples were positive for both of the quality-control primers, which indicated the quality of the extracted DNA was suitable for the intended PCR test purpose.
On the other hand, note that the single sample of postlarval L. vannamei was PCR positive for the five IHHNV primers applied, as was the IHHNV plasmid positive control sample. The L. vannamei sample was also positive for the two shrimp DNA quality-control primers, which target conservative DNA sequences in the genome of penaeid shrimp.
Inherited DNA
These PCR test data demonstrated that Moana monodon offspring derived from founder shrimp collected from the Indo-Pacific Region had genomic IHHN-related DNA sequences that reacted with the OIE-listed IHHNV diagnostic 392F/R and 389F/R primers. Therefore, caution is advised for the interpretation of conventional PCR test results for IHHNV when these primers are applied either singly or in tandem for the detection of IHHNV-infected P. monodon.
Also note the Table 1 data for the L. vannamei sample, which showed the expected PCR test reaction profile from a sample where IHHNV was present in the tissue. Note the positive PCR test results for all of the IHHNV PCR primers. This example showed the difference between shrimp tissue with IHHNV present and shrimp tissue with no virus, as demonstrated with the Moana monodon PCR test results.
The test outcome patterns were clear that where IHHNV was not present, the complete viral genome was not present. If the viral genome was not present, the virus itself was not present. Caution is advised in the interpretation of PCR test outcomes using primers that are known to occur naturally within the genome of monodon shrimp.
DNA inheritance
In order to pursue a hypothesis that the sequences were genomic in the monodon shrimp and not related to the presence of actual IHHNV, the authors carried out a series of studies to assess the inheritance pattern of the 392F/R DNA in offspring postlarvae spawned from individual male and female parent shrimp.
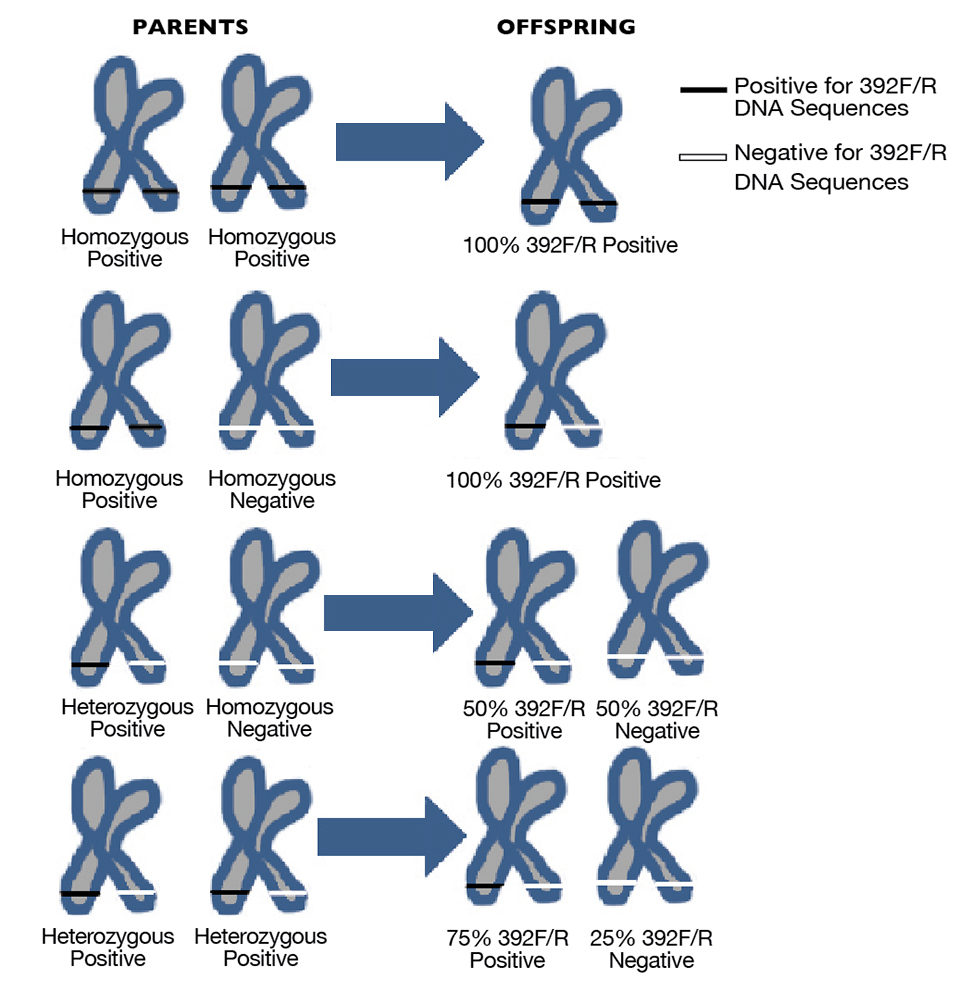
Individual postlarvae spawned from parents for which the 392F/R IHHN-related DNA sequences had been determined by PCR assay were sampled and tested. The hypothesis was that the 392F/R IHHN-related DNA sequences that occurred within at least one parent would be inherited in offspring shrimp, and the pattern of distribution in the offspring would be in accordance with Mendel’s Law of Segregation (Fig. 1).
Presented in Table 2 are the PCR test outcomes for three separate spawns from Moana breeders. The data show that the 392F/R IHHN-related sequences were inherited according to the Law of Segregation, and the DNA reacting with the PCR test primers must therefore have been within the genome of the parents. These DNA sequences were passed generationally from the parent to the offspring.
Brock, Examples demonstrate how 392F/R IHHN-related DNA sequences pass from parent to offspring, Table 2
| Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 |
|---|
Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 | Example 1 |
|---|---|---|---|---|---|---|---|---|---|
| Parents of P1 Family f6-xxx | SMP-DNA | 309F/R | 389F/R | 392F/R | Chi square statistic for f6-xxx. n = 43 | Chi square statistic for f6-xxx. n = 43 | Chi square statistic for f6-xxx. n = 43 | ||
| Female | Pos. | Neg. | Neg. | Neg. | Category | Observed | Expected | % Expected | Chi square value = 0.581 |
| Male | Pos. | Neg. | Neg. | Pos. | Positive | 24 | 21.5 | 50 | P = 0.45 |
| Negative | 19 | 21.5 | 50 | ||||||
| Example 2 | Example 2 | Example 2 | Example 2 | Example 2 | Example 2 | Example 2 | Example 2 | Example 2 | Example 2 |
| Parents of P1 Family f5-xxx | SMP-DNA | 309F/R | 389F/R | 392F/R | Chi square statistic for f5-xxx. n = 50 | Chi square statistic for f5-xxx. n = 50 | Chi square statistic for f5-xxx. n = 50 | ||
| Female | Pos. | Neg. | Neg. | Pos. | Category | Observed | Expected | % Expected | Chi square value = 0.286 |
| Male | Pos. | Neg. | Neg. | Pos. | Positive | 49 | 50.0 | 100 | P = N.A. |
| Negative | 1 | 0 | 0 | ||||||
| Example 3 | Example 3 | Example 3 | Example 3 | Example 3 | Example 3 | Example 3 | Example 3 | Example 3 | Example 3 |
| Parents of P1 Family f5-xxx | SMP-DNA | 309F/R | 389F/R | 392F/R | |||||
| Female | Pos. | Neg. | Neg. | Pos. | Category | Observed | Expected | % Expected | Chi square value = 0.286 |
| Male | Pos. | Neg. | Neg. | Pos. | Positive | 33 | 31.5 | 75 | P = 0.593 |
| Negative | 9 | 10.5 | 25 |
Sequencing analysis Moana has submitted samples of purified DNA amplified by 392F/R primers and excised from agarose gels for sequencing analysis to the Pacific Biosciences Research Center Biotech Core at the University of Hawaii at Manoa. Sequencing results received by e-mail are then further analyzed using the Basic Local Alignment Search Tool (BLAST).
The samples selected were from representative individual shrimp for the four groupings of genetic stock in the Moana monodon breeding herd. Tissue samples collected from 16 individual monodon shrimp and 16 different families were used to develop the findings presented in Table 3.
Brock, NCBI submission descriptions for IHHN-related genomic sequences, Table 3
| NCBI Blastn Accession Number | Description |
|---|
NCBI Blastn Accession Number | Description |
|---|---|
| EU675312.1 | IHHNV isolate Au2005 |
| EU675313.1 | IHHNV isolate Weipa (Northern Australia), 911 NS2 protein gene |
| AY124937.1 | IHHNV isolate from East Africa, non-structural protein 1 and protein genes |
| AY362547.1 | IHHNV from Thailand, non-structural proteins 1 and 2 |
| AY355307.1 | IHHNV isolate from Taiwan |
| AY590120.1 | IHHNV from Australia, non-structural protein 1 and 2 genes |
Note that for the Moana monodon with IHHN-related genomic DNA sequences, the Blastn accession number results were different from those of the tissue sample that had infectious IHHNV (Table 4). The sequence analysis findings further supported the conclusion that the DNA tested was very different between the monodon genomic DNA and the IHHNV DNA.
Brock, Submission descriptions for IHHNV sequences, Table 4
| NCBI Blastn Accession Number | Description, Origin |
|---|
NCBI Blastn Accession Number | Description, Origin |
|---|---|
| AF218266.2 | IHHNV complete genome |
| EF633688 | IHHNV isolate, Fujian |
| AY355308.1 | IHHNV isolate, Taiwan C |
| AY355306.1 | IHHNV isolate, Taiwan A |
| AY362548.1 | IHHNV from Ecuador, non-structural protein |
| AF273215.1 | IHHNV non-structural proteins 1 and 2 |
| AY590121.1 | IHHNV, New Caledonia |
Applications
In 2013, the presence of IHHN-related sequences in the genome of P. monodon has been public knowledge for the past seven years. Nevertheless, the authors believe that ongoing discussion about the IHHN-related DNA sequences in the genome of P. monodon can help keep people better informed about this irregularity and accept that a PCR-positive test using the 392F/R and 389F/R primers to assay for IHHNV does not by itself mean the shrimp were infected with IHHNV. As advised in the OIE Aquatic Manual, additional laboratory work is needed to clarify the source of the DNA that reacted positively in the PCR tests.
Moreover, the authors believe that the source location for the DNA sequences for a 392F/R positive PCR test reaction should not be based upon the application of the 389F/R PCR test, because their findings indicated that individual monodon shrimp can have genomic DNA sequences that prime up with these two PCR primer sets.
A second point is that when IHHNV is present in shrimp, all PCR primers typically react and display positive test results with the extracted DNA from the sample. This implies that when infectious IHHNV is present, DNA for the virus is also present.
However, when infectious IHHNV is not present, but shrimp have genomic DNA with the related sequences, positive PCR outcomes will not be found for the five PCR primer sets used. The simple explanation is that no “live” or infectious virus is present, because the virus genome is absent in the sample, and the entire virus genome must be present for an infectious and replicating virus to be present.
A third point is that there may be errors in the published literature regarding the classification of the different genomic types of infectious IHHNV, because the analysis work was carried out using tissue samples collected from P. monodon prior to learning that the presence of IHHN-related DNA sequences were part of the genomic DNA of P. monodon. Thus, if two sources (virus and host) contributed to the extracted DNA that made up the analyzed samples, it would be unreliable to assume the origin of the DNA for the sequence generated in the PCR test was only the IHHNV.
(Editor’s Note: This article was originally published in the September/October 2013 print edition of the Global Aquaculture Advocate.)
Authors
-
J.A. Brock, DVM
Moana Technologies, LLC
99-1255 Waiua Place #4
Aiea, Hawaii 96701 USA -
Nathene Antonio
Moana Technologies, LLC
99-1255 Waiua Place #4
Aiea, Hawaii, USA -
Brad Argue, Ph.D.
Geovanni Tolentino
Moana Technologies, LLC
Kailua-Kona, Hawaii, USA
Tagged With
Related Posts

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
AHPN inferences based on behavior of vibrio bacteria
Vibrio parahaemolyticus, a strain of which is the cause of acute hepatopancreatic necrosis (AHPN), has both virulent and benign strains. This strain colonizes the stomachs of shrimp by the formation of a biofilm, which protects it from antibiotics and other potential treatments.

Health & Welfare
Measuring susceptibility differences to IHHNV infection
This study evaluated the susceptibility to IHHNV in three different shrimp batches by measuring infectivity titers. Each batch showed a different infectivity titer, therefore, each batch had a specific susceptibility to IHHNV.

Health & Welfare
Animal health giants have sea lice in their crosshairs
Alltech and Benchmark have been working on the next generation of sea lice solutions and believe they have new products that can help salmon farmers win.


