Virus has caused severe mortalities in some shrimp, diminished effects in others
The genomic sequences of viruses change through time, either from random mutation or selection pressure. This leads to the formation of virus strains that can vary considerably in their effects on host organisms. Many examples are found in human or animal viruses. For example, strains of influenza virus differ in virulence, and a vaccine that is effective against one strain may not be against another.
Although similar variation may be expected in shrimp viruses, little information is available on the genetic variation of shrimp viruses or its significance.
IHHNV
The University of Arizona Aquaculture Pathology Laboratory in Tucson, Arizona, USA, receives samples of diseased shrimp from around the world and maintains an archive of tissue samples from viral-infected shrimp. This has afforded the opportunity to study genetic variation among isolates collected over a wide geographic area.
Of particular interest is the Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV). This virus is widespread and has been detected in stocks around the world, including the Americas, Oceania, Africa, and Asia. Its effects vary substantially among susceptible host species, and its infection in at least one species appears to have changed.
IHHNV has caused severe mortalities in some shrimp species but diminished effects in others. In 1981, when the virus was first discovered in (Penaeus stylirostris), it was associated with mortalities of up to 90 percent in infected populations in Hawaii. In more recent cases of IHHNV infection in P. stylirostris, the virus appears to be less virulent, suggesting either a change in the virus or a change in the defenses of the host.
Other species of penaeid shrimp respond differently to IHHNV infection. For example, IHHNV is not known to cause significant mortalities in P. vannamei or P. monodon, but it can result in a disease known as Runt Deformity Syndrome, which greatly reduces growth and may cause cuticular and rostrum deformities.
Analyses of variation
The authors recently studied samples of IHHNV-infected P. stylirostris, P. vannamei and P. monodon from a wide range of geographic areas including Hawaii, USA; Mexico; Colombia; Ecuador; Panama; the Philippines; Thailand; Taiwan; Madagascar; and Tanzania.
Three approaches were taken in an analysis of the variations among isolates. The first compared the IHHNV genome sequences from samples collected from different locations, the second compared the histopathological changes in the infected shrimp, and the third determined the effects of the viral isolates on shrimp infected in laboratory bioassays.
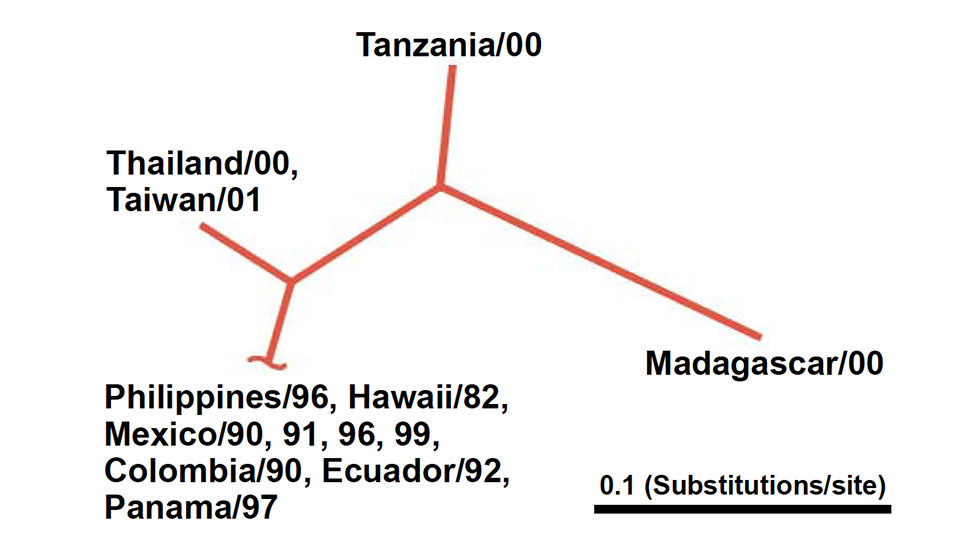
Sequence variation
For each sample, the nucleic acid sequence of a selected region of the viral genome was determined. These were then compared through the use of statistical clustering methods developed for phylogenetic analysis.
Results revealed four groups based on DNA sequence identity. The Philippine isolate had a 99.8 percent nucleotide sequence identity with isolates from Hawaii and Central/South America. At 96.2 percent, the Thailand isolate showed a slightly lower identity with the Hawaii isolate, but was virtually identical to the Taiwan isolate. The isolates from Tanzania and Madagascar differed the most (8-14 percent) from the other samples.
Histopathology
Shrimp infected with these isolates were analyzed for the presence of Cowdry type A intranuclear inclusion bodies (CAIs) in the principal target tissues by routine histological methods. The hallmark CAIs were found in the shrimp infected with IHHNV isolates from Hawaii, the Philippines, and Thailand, indicating that the small 4 percent difference in DNA sequence did not change the characteristic histopathology of the virus.
These infected shrimp were also shown to be positive for IHHNV infection by in situ hybridization. However, the CAIs were not found in shrimp exposed to the two Africa isolates, and these samples were negative for IHHNV with in situ hybridization.
Bioassays
For the Philippines and Thailand IHHNV isolates, laboratory infections were carried out in bioassays using specific pathogen-free P. vannamei as indicator shrimp. In these trials, there were no mortalities among the indicator shrimp one month after exposure to IHHNV. The challenged shrimp tested positive for IHHNV with PCR and in situ hybridization.
In a challenge study with Madagascar IHHNV, the shrimp showed no mortality after one month. In addition, both PCR and in situ hybridization were negative for IHHNV, indicating the Madagascar IHHNV was not infectious to P. vannamei.
Results
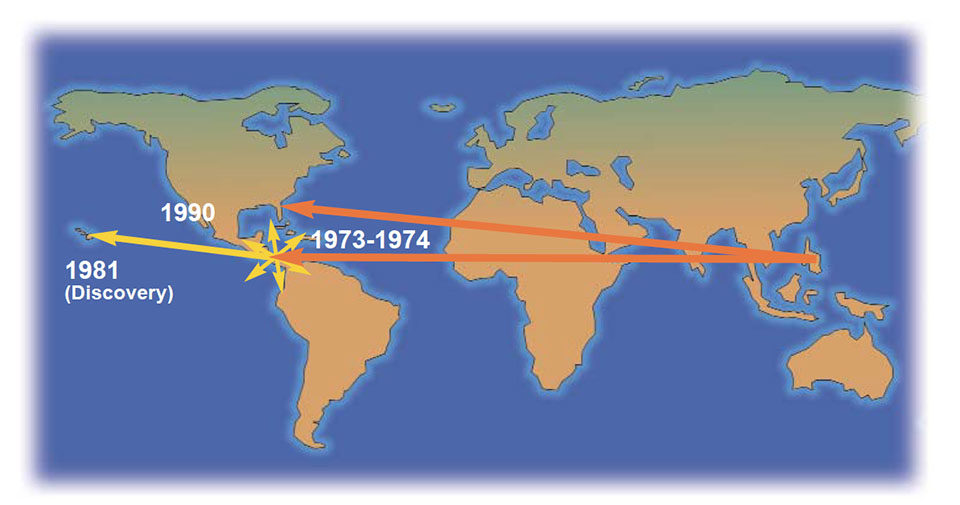
This information allows piecing together the puzzle of how this virus has spread. The data on sequence analysis supports the theory that the Philippines was the original source of IHHNV. It was introduced through P. monodon stocks into the Americas in the 1970s and was later “discovered” in P. stylirostris cultured in Hawaii. Likewise, the high similarity between the isolates from Taiwan and Thailand likely resulted from the frequent transfer and importation of live broodstock shrimp from Thailand to Taiwan.
The nature of the two Africa IHHNV isolates is not clear. They were the most divergent isolates with regard to DNA sequence. The Africa isolates were detected with PCR but not with histology or in situ hybridization.
In the bioassays, infections with Madagascar isolate were not achieved in either P. monodon or P. vannamei. There are a number of possibilities for these results. It is possible the level of IHHNV infection in African P. monodon was very low and therefore could not be detected by histology and in situ hybridization. Alternatively, the virus may be present in a form that is no longer infectious. The sequence variation in the genomic region responsible for replication was higher than that found in the other IHHNV isolates and most parvoviruses in general.
This may indicate the isolates were no longer capable of replicating and that the remnant of the IHHNV genome was associated with the P. monodon host. If so, this could explain why IHHNV in west Indian Ocean P. monodon appears not to be infectious.
Conclusion
Nucleic acid sequence data can be used to identify the geographic origins of viruses that infect penaeid shrimp. A study of sequence variation revealed considerable differences among IHHNV strains and their pathological effects. In addition, the work suggested how at least two strains of the virus became so widespread.
The emergence of IHHNV in the Western Hemisphere three decades ago may have been from introductions of IHHNV-infected P. monodon from the Philippines. Because IHHNV infection is asymptomatic in P. monodon except in severe cases, the presence of the virus may have escaped notice until stocks of other, more susceptible species became infected.
(Editor’s Note: This article was originally published in the February 2004 print edition of the Global Aquaculture Advocate.)
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-

Kathy Tang
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, AZ 85721 USA -

Donald Lightner, Ph.D.
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, AZ 85721 USA
Tagged With
Related Posts

Aquafeeds
Biosecurity protocols needed for shrimp feeds, feeding practices
Shrimp aquafeeds – live, fresh or formulated – should not be an entry point of potential pathogens to the shrimp and/or to their culture systems.

Health & Welfare
AHPN inferences based on behavior of vibrio bacteria
Vibrio parahaemolyticus, a strain of which is the cause of acute hepatopancreatic necrosis (AHPN), has both virulent and benign strains. This strain colonizes the stomachs of shrimp by the formation of a biofilm, which protects it from antibiotics and other potential treatments.

Health & Welfare
Big shoes to fill: Dhar takes reins at shrimp pathology laboratory
Arun Dhar, Ph.D. will attempt to fill the “big shoes” of Dr. Donald Lightner at the University of Arizona’s Aquaculture Pathology Laboratory, where the shrimp disease EMS was diagnosed.

Health & Welfare
Genomic IHHN-related DNA sequences in black tiger shrimp
Polymerase chain reaction (PCR) testing demonstrated that Penaeus monodon offspring derived from founder shrimp collected from the Indo-Pacific region had genomic infectious hypodermal and hematopoietic necrosis-related DNA sequences that reacted with OIE-listed IHHN virus diagnostic primers.

