RT-LAMP-NALF method is specific, sensitive and user-friendly

Infectious myonecrosis disease is currently a crustacean disease notifiable to the World Organisation for Animal Health. Despite the availability of rapid and accurate detection methods for infectious myonecrosis virus (IMNV), the virus that causes the disease, delays and misdiagnosis of infectious shrimp diseases are common in marine shrimp culture zones because the equipment necessary for pathogen detection is beyond what most diagnostic sites can afford. The authors therefore worked to advance simple, specific and sensitive diagnostic tests to diagnose infectious myonecrosis.
With grant support from the U.S. Consortium Marine Shrimp Farming Program; Cooperative State Research, Education and Extension Service; U.S. Department of Agriculture; National Council for Scientific and Technological Development; Brazil Ministry of Science and Technology and National Fisheries Institute, they developed a reverse transcription loop-mediated isothermal amplification and nucleic acid lateral flow (RT-LAMP-NALF) assay for the detection of infectious myonecrosis in resource-poor diagnostic settings.
Procedure development
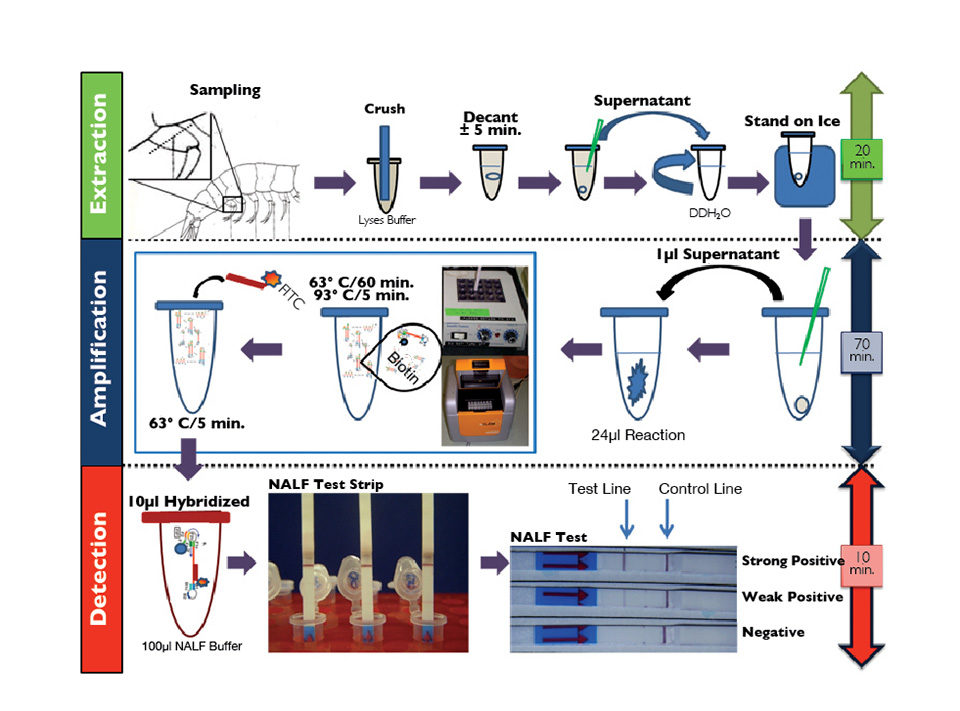
The RT-LAMP-NALF method combines simplified nucleic acid extraction, a reverse-transcription isothermal amplification platform and one-step visual colorimetric confirmation of the IMNV amplified sequences using a generic NALF qualitative detection test strip (Fig. 1).
IMNV-specific RT-LAMP primers were designed for detecting the structural capsid protein gene of IMNV. For purposes of comparison, efforts were made to design primers that amplify the 365-to 568-bp region, which encompasses the 412- to 545-bp region amplified by real-time reverse transcription-polymerase chain reaction (RT-PCR).
The sensitivity of RT-LAMP using two or three primer pairs and nested RT-LAMP using three primer pairs was compared by real-time RT-PCR with a TaqMan probe. IMNV isolates were obtained from the archives of diagnostic materials collected from 2003 to 2008 at the Aquaculture Pathology Laboratory of the University of Arizona.
Sensitivity, specificity
The RT-LAMP using three primer pairs was 10-fold more sensitive than when using two primer pairs (10-4), and the 10-3 and 10-2 dilutions were more intensely amplified by RT-LAMP when using three rather than two primer pairs. The addition of the third primer pair to the reaction tube increased sensitivity and specificity – a pattern of results also obtained in other assays.
The detection of IMNV RT-LAMP products was accomplished using an NALF test strip. The biotin-labeled IMNV RT-LAMP amplicons were hybridized with an FITC-labeled probe specific to the IMNV capsid gene. The hybridization product was dotted for sample application of the NALF strip, the IMNV RT-LAMP immune complexes were captured, and a red test band was generated. Labeled immune complexes not captured on the test line overflowed the control band and were immobilized by species-specific antibodies, resulting in an intense control band.
The RT-LAMP-NALF showed equivalent sensitivity to RT-LAMP with three primer pairs. On one hand, the RT-LAMP-NALF was found to be 100 and 10 times more sensitive than one-step RT-PCR and RT-LAMP with two primer pairs, respectively. On the other hand, the RT-LAMP-NALF was 10 and 100 times less sensitive than nested RT-PCR and real-time RT-PCR, respectively.
The specificity of the RT-LAMP-NALF method did not react with hepatopancreatic necrosis virus, white spot syndrome virus, infectious hypodermal and haematopoietic necrosis virus, yellow head virus, Taura syndrome virus or Penaeus vannamei nodavirus.
Andrade, Results of specificity from assays using real-time RT-PCR and RT-LAMP-NALF for IMNV, Table 1
| Isolate | Year | Real-Time RT-PCR (IMNV copies/µl RNA) | RT-LAMP-NALF |
|---|
Isolate | Year | Real-Time RT-PCR (IMNV copies/µl RNA) | RT-LAMP-NALF |
|---|---|---|---|
| 1 | 2003 | 8.78 x 104 | + |
| 2 | 2003 | 5.99 x 104 | + |
| 3 | 2004 | 2.33 x 104 | + |
| 4 | 2005 | 3.44 x 104 | + |
| 5 | 2005 | 4.42 x 107 | ++ |
| 6 | 2006 | 5.34 x 106 | ++ |
| 7 | 2006 | 7.96 x 105 | ++ |
| 8 | 2006 | 1.99 x 106 | ++ |
| 9 | 2006 | 3.28 x 106 | ++ |
| 10 | 2006 | 9.19 x 106 | ++ |
| 11 | 2007 | 3.92 x 105 | ++ |
| 12 | 2007 | 4.48 x 106 | ++ |
| 13 | 2008 | 3.15 x 104 | + |
+ = Weakly positive, ++ = Strongly positive.
Effective testing
These results demonstrated that the RT-LAMP-NALF method was specific, sensitive and user-friendly. The RT-LAMP-NALF method also can shorten the time for analysis and has potential application for IMNV diagnosis in resource-poor and point-of-care diagnostic settings, because it does not require expensive, specialized equipment and carcinogenic ethidium bromide.
(Editor’s Note: This article was originally published in the July/August 2009 print edition of the Global Aquaculture Advocate.)
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Author
-
Thales P.D. Andrade
University of Arizona
Department of Veterinary Science and Microbiology
Aquaculture Pathology Laboratory
Tucson, Arizona 85721 USA
Tagged With
Related Posts

Health & Welfare
A comprehensive look at the Proficiency Test for farmed shrimp
The University of Arizona Aquaculture Pathology Laboratory has carried out the Proficiency Test (PT) since 2005, with 300-plus diagnostic laboratories participating while improving their capabilities in the diagnosis of several shrimp pathogens.

Responsibility
A look at various intensive shrimp farming systems in Asia
The impact of diseases led some Asian shrimp farming countries to develop biofloc and recirculation aquaculture system (RAS) production technologies. Treating incoming water for culture operations and wastewater treatment are biosecurity measures for disease prevention and control.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
Brazil shrimp farm performs genetic selection for IMNV resistance, growth
The Queiroz Galvão Alimentos shrimp farm and hatchery in Brazil have been working with Concepto Azul to implement a disease-prevention and genetic-breeding program that addresses ongoing impacts from infectious myonecrosis virus (IMNV) and other pathogens.


