Quantification of pathogen in infected animals crucial for monitoring disease progression

Acute hepatopancreatic necrosis disease (AHPND), also known as early mortality syndrome, has caused severe mortalities in farmed populations of Litopenaeus vannamei and Penaeus monodon shrimp. The disease has led to significant production and economic losses at shrimp farms and to the aquaculture industry in general in affected regions.
Clinical signs of the disease in infected shrimp include an empty gastrointestinal tract and whitish, atrophied stomach and hepatopancreas. The causative agent was determined to be the unique strains of the bacterium Vibrio parahaemolyticus that contain the pirA- and pirB-like toxin genes. These genes are located in a large, 69-kb plasmid. Plasmids are self-replicating, double-stranded DNA molecules that pass between bacterial cells through conjugation and transformation.
Initially, diagnosis of AHPND could only be accomplished through histological examinations and laboratory bioassays. Later, molecular diagnostic methods based on conventional polymerase chain reaction testing targeting pirA- and/or pirB-like genes became available. However, quantification of the AHPND pathogen in the infected animals is one of the most important means for monitoring disease progression.
Bacterial quantities can be determined through counting colony-forming units (CFUs), but this is a laborious and time-consuming method. Real-time, quantitative polymerase chain reaction (qPCR) analysis has become an attractive alternative for its advantages of speed, sensitivity and specificity.
PCR primers, probe
The PCR primers and specificity-enhancing hydrolysis probe for the detection of the virulence plasmid were selected from the pirA-like gene. The primers were used to amplify a DNA fragment of 135-bp. The probe was synthesized and labeled with different dyes on either end.
Samples of extracted DNA were added to a qPCR mixture containing 0.3 µ of each primer and 0.1 µ probe to a final volume of 10 µL. The qPCR profile consisted of 20 seconds at 95 degrees C, followed by 40 cycles of 3 seconds at 95 degrees C and 30 seconds at 60 degrees C.
Sensitivity, specificity
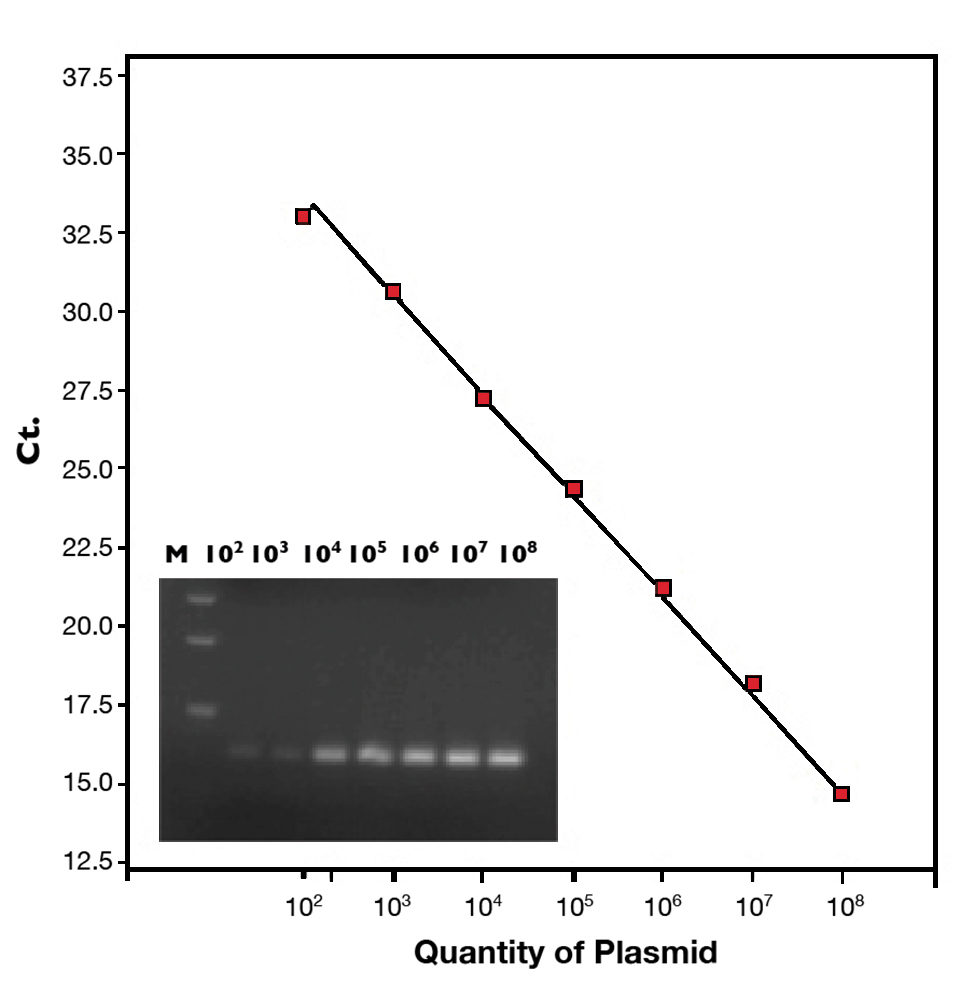
The detection limit of this qPCR assay was less than 10 copies of virulence plasmid. The standard curve (102 to 108 copies of plasmid) is shown in Figure 1. This method can detect 12 AHPND-causing V. parahaemolyticus isolates from Mexico and Vietnam, but did not cross react with 35 non-pathogenic Vibrio bacteria.
Affect on farmed shrimp
The authors used two cases of farm-raised L. vannamei collected in 2012 to detect and quantify the AHPND plasmid. Case 1 consisted of 32 small juvenile shrimp collected from China that showed significant mortalities and severe effects of AHPND, determined by histological examination.
By qPCR, DNA extracted from each shrimp proved to be AHPND-positive. The quantities of virulence plasmid ranged from 2.5 x 103 to 4.7 x 106 copies per milligram of tissue (Table 1).
| Laboratory Bioassay | Infection Route | Shrimp | Copies/mg Tissue |
|---|---|---|---|
| 1 | Per os (1 x 109 CFU/ml) | Day 1 (moribund/dead) Day 2 (moribund/dead) | 2.5 x 103-4.7 x 106 2.1 x 105-3.6 x 106 |
| 2 | Immersion (2 x 105 CFU/ml) | Day 1 (moribund/dead) Day 2 (moribund/dead) Day 9 (surviving) | 1.8 x 103-2.1 x 105 1.4 x 105-7.9 x 105 |
| 3 | Per os (8 x 108 CFU/mL) | 1.8 x 101-3.9 x 102 | |
| 4 | Per os (1 x 109 CFU/mL) | Water sample | 3.5 x 102-2.2 x 106* |
The second case included 12 juvenile shrimp collected from the Ben Tre Province of Vietnam. These shrimp suffered high mortalities and were later found to have AHPND through histological examination. In qPCR analysis, DNA extracted from hepatopancreas tissues found five of the 12 shrimp were positive for AHPND. The quantities of virulence plasmid were 3.9 to 5.8 x 105 copies per milligram of tissue.
Virulence plasmid copy numbers varied widely, ranging 4-105-6/mg tissue. Thus, the AHPND bacterial load in affected shrimp would be expected to also be highly variable. This suggested that factors other than bacterial load, such as shrimp age or size, the presence of other pathogens or environmental conditions, may influence the mortalities in affected farms.
Laboratory-infected shrimp
Shrimp collected from laboratory infections were analyzed for AHPND. In bioassay 1, a per os feeding experiment, specific-pathogen-free L. vannamei with a mean individual weight of 1 g were fed shrimp feed mixed with AHPND V. parahaemolyticus. The bacteria were first grown to 1 x 109 CFU/mL and mixed with shrimp feed at a 1:1 ratio.
All 50 infected shrimp became moribund or died within two days. These shrimp were positive for AHPND by histological examination and qPCR. The quantities of virulence plasmid ranged from 2.5 x 103 to 4.7 x 106 per milligram of tissue (Table 1).
In bioassay 2, shrimp were exposed to AHPND V. parahaemolyticus by immersion at a concentration of 2 x 105 CFU/mL water. All the shrimp died within two days. By qPCR, the quantities of virulence plasmid in the moribund/dead shrimp ranged from 1.8 x 103 to 7.9 x 105 copies per milligram of tissue (Table 1).
Bioassay 3 was a challenge study to a selected line of L. vannamei. At a mean weight of 2 g, the shrimp were stocked in 1,000-L tanks and given feed containing AHPND V. parahaemolyticus. At day 9, 244 shrimp (48 percent) survived, whereas none of the 50 shrimp in the positive control tank survived after two days.
Histological examination did not detect AHPND in these surviving shrimp. By qPCR, the quantities of AHPND plasmid were 18-390 copies/mg tissue (Table 1), suggesting this selected line was better at fending off the disease. However, these surviving shrimp, which carried low quantities of pathogenic bacteria, could act as vectors capable of spreading the disease.
Although hepatopancreas tissue is usually sampled for detecting AHPND V. parahaemolyticus, a fourth bioassay was performed to demonstrate that water samples can be used for AHPND diagnostics.
In this bioassay with shrimp fed feed mixed with AHPND V. parahaemolyticus, water was sampled from each tank at day 5 and concentrated 50 times. AHPND was detected in seven of nine water samples by the highly sensitive qPCR, ranging 3.5 x 102 to 2.2 x 106 plasmid copies/mL water (Table 1). This is relevant, since it has been reported that AHPND can be transmitted via water from AHPND-affected ponds.
(Editor’s Note: This article was originally published in the May/June 2015 print edition of the Global Aquaculture Advocate.)
Authors
-

Jee Eun Han, DVM, Ph.D.
School of Animal and Comparative Biomedical Sciences
University of Arizona
Tucson, Arizona 85721 USA[117,100,101,46,97,110,111,122,105,114,97,46,108,105,97,109,101,64,110,97,104,101,101,106]
-

Kathy Tang, Ph.D.
School of Animal and Comparative Biomedical Sciences
University of Arizona -
Brenda White
School of Animal and Comparative Biomedical Sciences
University of Arizona -

Donald Lightner, Ph.D.
School of Animal and Comparative Biomedical Sciences
University of Arizona
Tagged With
Related Posts

Health & Welfare
Rapid on-site diagnostic tool for AHPND management
To help minimize economic losses caused by acute hepatopancreatic necrosis disease, timely detection of its pathogenic Vibrio etiological agent plays a critical part in disease management. A portable device with accompanying polymerase chain reaction assays can quickly identify the AHPND markers in shrimp, water, live feed or other sources.

Health & Welfare
Four AHPND strains identified on Latin American shrimp farms
Two virulence genes are known to encode a binary photorhabdus insect-related toxin that causes acute hepatopancreatic necrosis disease in shrimp. The pathogenicities of these V. campbellii strains were evaluated through laboratory infection and subsequent histological examination in P. vannamei shrimp.

Health & Welfare
PCR methods characterize AHPND V.p isolates
In analyzing the plasmid sequence from the whole genome sequences of AHPND V. parahaemolyticus (V.p) isolates, researchers identified a clear geographical variation within the plasmid, and developed PCR methods to characterize AHPND V.p isolates as either Mexico-type or Southeast Asia-type.

Health & Welfare
A holistic management approach to EMS
Early Mortality Syndrome has devastated farmed shrimp in Asia and Latin America. With better understanding of the pathogen and the development and improvement of novel strategies, shrimp farmers are now able to better manage the disease.


