Technology has improved the accuracy and affordability of microbiome analysis
From boiling hydrothermal vents of our deep oceans, extremely cold polar environments, Yellowstone National Park’s hot springs, to both animals’ and our own intestines, microbial communities inhabit virtually any location on this planet. Largely invisible to the naked eye, microbes contribute approximately 4-6 × 1030 cells to Earth’s biosphere, or 2 to 5 orders of magnitude more than the total number of animal and plant cells.
Several hundred to a thousand times smaller than fine sand particles, microorganisms are comprised of viruses, fungi, protists, archaea and most notably bacteria that shape our ecology through their collective metabolic input. Generally speaking, microbes that have adapted to their environment will thrive in that niche, outcompeting those microbes that may not be as well suited. Not all microbial communities inhabiting various ecosystems are alike.
The nature of their surrounding physiochemical factors helps shape the complexity of the consortium and their role in making the ecosystem function. Nevertheless, in most environments, the true function of these distinct microbial communities – including aquaculture systems – has yet to be fully understood.
Microbial communities have diverse roles
Recent studies have shown that animals, including humans, rely on their microbial communities for the crucial role these microbes play in supporting their host’s health. For example, certain species of Escherichia coli have been acknowledged as happy symbionts in the gut, benefiting from the nourishment of the daily human diet, and repaying their host by producing essential vitamins which are absorbed in our intestines.
In fact, the role of host-friendly bacteria has been a topic of interest in the scientific community, and research has described the many benefits of certain microbial communities in training the immune system, aiding in food digestion, preventing the invasion of harmful bacteria, and contributing to the overall health of the organism.
On the other hand, chaos in the microbial consortium may result in adverse effects to their host’s health. Major shifts in an organism’s typical microbial community structure may be brought on by environmental pressures, nutrient availability, chemical exposure, or invasion of harmful microbial species. In some situations, these shifts may be benign, and normal “homeostatic” population may return. In other cases, these disruptions may have severe long-term consequences to the host.
Beneficial host-microbe relationships extend to all animals, and particularly to those of agricultural importance, where understanding the role of bacteria to their host can be highly beneficial to producing quality supply. Like humans, cows and other livestock rely on their gut bacterial communities for efficient digestion, thus signifying the importance of maintaining healthy gut bacterial profiles as necessary for nutrient uptake.
In fact, efforts to understand the normally occurring microbial populations in production and companion animals has led to practical applications, such as the focused use of microbes as probiotics to provide health benefits while minimally disrupting the naturally occurring microbial residents. Current applications of probiotics have shown success, by way of: 1) stabilizing intestine microbial communities, (2) reducing the onset of gut-related morbidities without antibiotics, (3) promoting weight gain, and (4) reducing waste production.
Microbiome analysis and aquaculture
Aquaculture practices have expanded since the early 1950s, with a global projection of over 60 percent of edible fish to be a result of aquaculture systems by 2030. With this expansion, it has become advantageous to make full use of the controlled environment offered by aquaculture. For improving organism health and quality supply, this regulation has prompted prebiotic feeding regiments, controlled breeding, feed nutrient optimization and reducing disease by antibiotics.
This has also included developing strategies for clean recirculating water in closed systems and offering a venue for scientific breakthroughs and discovery – all while reducing the largely overfished wild stocks, coastal water pollution and anthropogenic methane production. Even with recent leaps in improving fish health and protein quality in aquaculture, we still do not fully understand, nor have we harnessed the associated microbes for their proven benefits to health.
Whether finfish or invertebrate, salt or freshwater, all aquaculture species will have a co-dependent population of microbes that, in many cases, will determine the success of production for those species. Unfortunately, the world of the microbiome has been largely ignored, and until now, the 1 trillion-plus living bodies in the human gut alone, along with their potential role in host function, have been overlooked.
Powerful new technologies
Fortunately for aquaculture, the new millennium brought with it powerful technologies that have allowed researchers to pry into the world of microbial communities as they harmoniously co-exist with other organisms. Specifically, NextGen DNA sequencing alongside powerful bioinformatics computer software has contributed to the study of the microbiome, the term given to the collective DNA of microorganisms (microbiota) from a niche. Application of this technology has made it possible to infer identities, functions and population trends based solely on ubiquitously sequencing microbial DNA, sparking numerous efforts to deconstruct how microbes contribute to host health and disease.
As this technology has become more available and affordable over time, the past few years have seen a spark in microbiome studies focusing on popular aquaculture organisms. To date, several studies have made significant headway in defining: 1) what a healthy microbial profile looks like, 2) how these communities can be disrupted, 3) what the consequences are of these disruptions, and 4) what specific microbial species or core microbial community contribute most to host health or disease.
The acquired knowledge in these areas of interest have opened a door: unlocking the potential to monitor, maintain, modulate and harvest microbial communities, specifically in aquaculture species. As the aquaculture industry continues to grow, awareness of this invisible world may hold the key to promote health, increase nutrient uptake, stave off disease and revolutionize our approach to producing and sustaining quality supply.
Knowledge of microbes enhances aquaculture performance
The real question is this: Can knowledge of microbes lead to enhancing aquatic animal health and production at a time when enhanced food production is required worldwide? The answer lies with the state of current research and future explorations by scientists and aquaculturists alike. In recent years, technology has improved the accuracy and affordability of microbiome analysis and continues to be a more user-friendly platform. Introducing such approaches to aquaculture could clarify the microbial profiles unique to the specific organisms in relation to their aquaculture environment.
Understanding the stable microbiota of cultured aquatic organisms could enhance our ability to monitor those communities to ensure health, good nutrition and predict likely disease states. We would predict that the microbial communities of fish are diverse among different fish species, so the more information that is shared with the aquaculture community on microbiota, the more likely we are to coalesce on those microbial species or populations that are likely to contribute to host nutrition and health.
Perspectives
Focusing on preserving and promoting those innate bacterial species that provide health benefits could lead to transformative results, from maximizing nutrient uptake to producing high protein yields and improving the fish immune system to fight diseases. This will extend to devising species-specific probiotic regiments.
Formulating microbial probiotics may forego the dependence on antibiotic treatments altogether. This can reduce antibiotic leaks into the environment, counterbalance emergent antibiotic resistant bacteria and lead to conscientious exportation of stock.
At the very least, coupling broad-spectrum antibiotics with specific probiotic bacteria additives could reduce the incidence of antibiotic related side-effects in fish populations. Considering how water treatments (previously thought to be innocuous) could throw off fish microbes in aquaculture would be crucial in maintaining large-scale fish health.
In addition, let’s not limit ourselves to just finfish. There are many species without a backbone whose culture could be enhanced by understanding the role of microbiota in their growth and health.
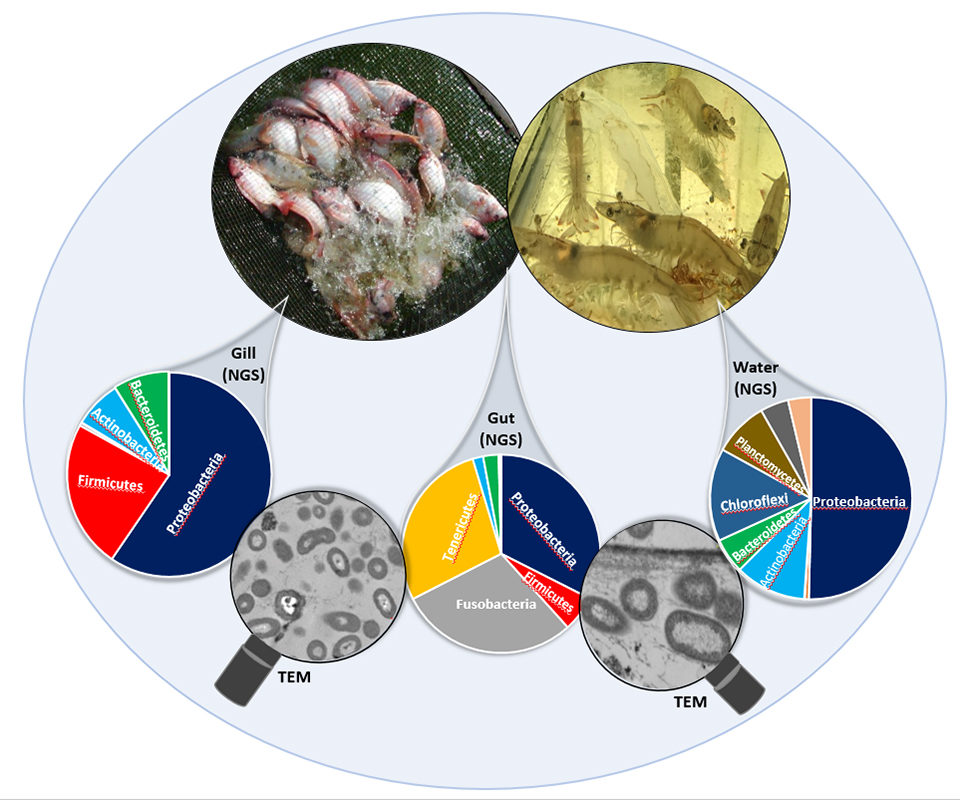
Lastly, determining the diverse roles of microbial populations across the organism, from the gut to the gills, could unlock a trove of information leading to innovative approaches to promote health through microbiota. Understanding this hidden world could unlock a potential that may change the practice of aquaculture altogether.
Acknowledgements: Research supported in part by the Nutrition and Obesity Research Aquatic Animal Research Core (P30DK056336) and Microbiome Resource at UAB School of Medicine.
Authors
-
Joseph A. Hakim
Ph.D. Candidate
Department of Biology
The University of Alabama at Birmingham
Birmingham, Alabama 35294 USA -
Hyunmin Koo
Ph.D. Candidate
Department of Biology
The University of Alabama at Birmingham
Birmingham, Alabama 35294 USA -
Stephen A. Watts
Corresponding author
Professor
Department of Biology
The University of Alabama at Birmingham
Birmingham, Alabama 35294 USA
Tagged With
Related Posts

Aquafeeds
A new nutrient for aquaculture, from microbes that consume carbon waste
Biotechnology firm NovoNutrients aims to produce a line of nutraceutical aquafeed additives as well as a bulk feed ingredient that can supplement fishmeal. Its process includes feeding carbon dioxide from industrial gas to a “microbial consortium” starring hydrogen-oxidizing bacteria.

Health & Welfare
Ammonia addition enhances microbial flocs in nursery phase for Pacific white shrimp
In a study, “pre-fertilization” in the nursery phase of a biofloc system for shrimp was tested. The objective was to accelerate the biofloc formation to minimize ammonia concentrations, avoiding high peaks during culture.

Aquafeeds
KnipBio partners with ICM to scale-up production of single-cell protein for aquaculture
Leader in fermentation, bio-refining technology a “great strategic fit” for single-cell protein aquafeed ingredient maker transitioning from R&D labs to commercial production.

Innovation & Investment
Algae innovators aim to freeze out early-stage shrimp losses
A greenhouse in Belgium believes its innovative shrimp feed product, made from freeze-dried microalgae, packs the necessary nutrients for the crustacean’s most vulnerable life stage: the first three days of its life.


