Genotypes from four wild populations compared to genetic profiles of captive stocks
The implementation of animal breeding programs is enacted through managed inbreeding for selective traits of economic importance. In the process, there will be unintentional diversity loss in the genetic profiles of the broodstock from generation to generation. Such loss can be tracked with the use of microsatellite markers in as little as two generations.
Genetic masking evolves when parent stocks from different populations are crossbred to establish new families. Fast forward several generations, and many programs lose the ability to track data from the parent set representative of the original population.
Specific breeding strategies are required in most captive programs to prevent or minimize genetic loss over multiple generations. The rate of loss for genetic diversity is amplified intentionally when the population is subjected to selection for trait improvement, and unintentionally when there is no breeding strategy for minimizing the loss in captive maintenance of most small breeding populations.
Study setup
The authors conducted studies to determine the extent to which farmed populations of Pacific white shrimp (Litopenaeus vannamei) showed loss of genetic diversity relative to their wild founding stocks, and whether the founding stocks could be identified from the genetic makeup of the farmed animals. Research also examined whether genetic markers could be used to identify farmed animals and provide stock protection for breeders who develop specialized lines of animals.
Genotypes from four wild populations were compared to the genetic profiles of captive stocks from Ecuador, Panama, and Mexico. The shrimp from Panama were known to have originated from Panamanian wild stocks. For Ecuador, the captive stocks were several generations removed from their original Panamanian wild stocks. Although of uncertain origin, the captive Mexican stocks reportedly came from Venezuela and were compared to Mexican wild stocks. The diversity of all the captive stocks was compared to that of wild stocks of origin once confirmed by genotype assignment.
Results
All captive stocks showed varying losses of diversity, but could be associated with their originating founding stocks. Table 1 shows the loss of diversity of the farmed animals compared to the wild stocks from four locations.
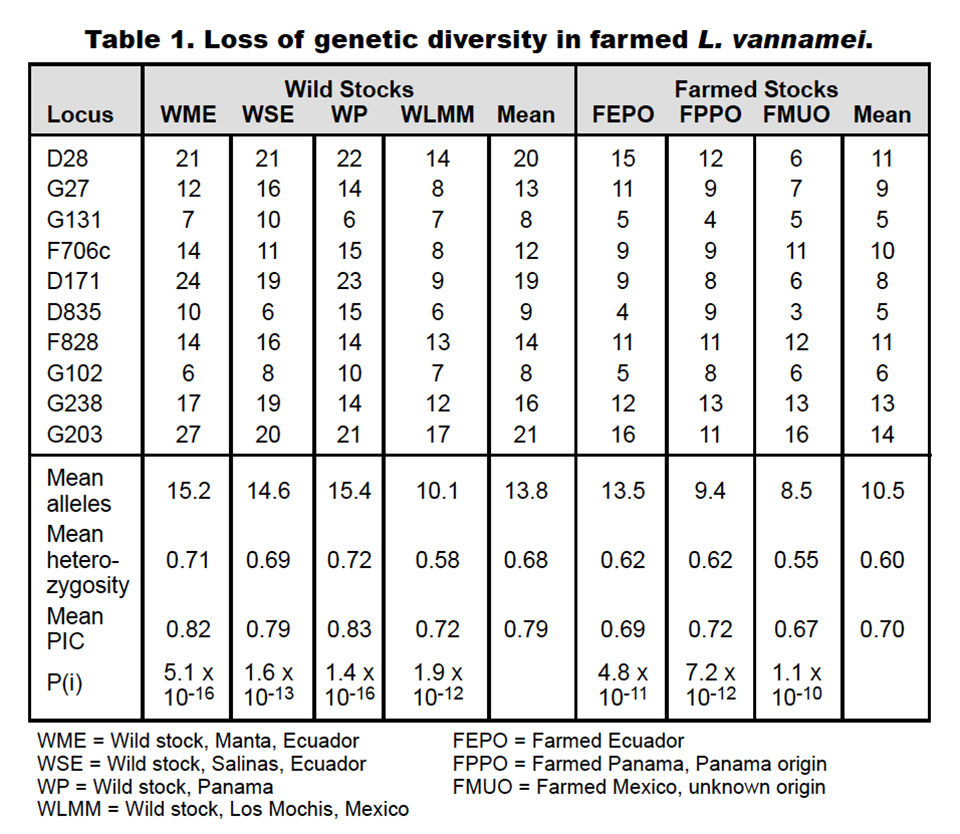
The founding stocks of two of the farmed populations (FEPO and FPPO) were known prior to the comparison, while the third (FMU) was not. Loss of diversity was based on the reduction in number of alleles, heterozygosity, polymorphic information content and other factors.
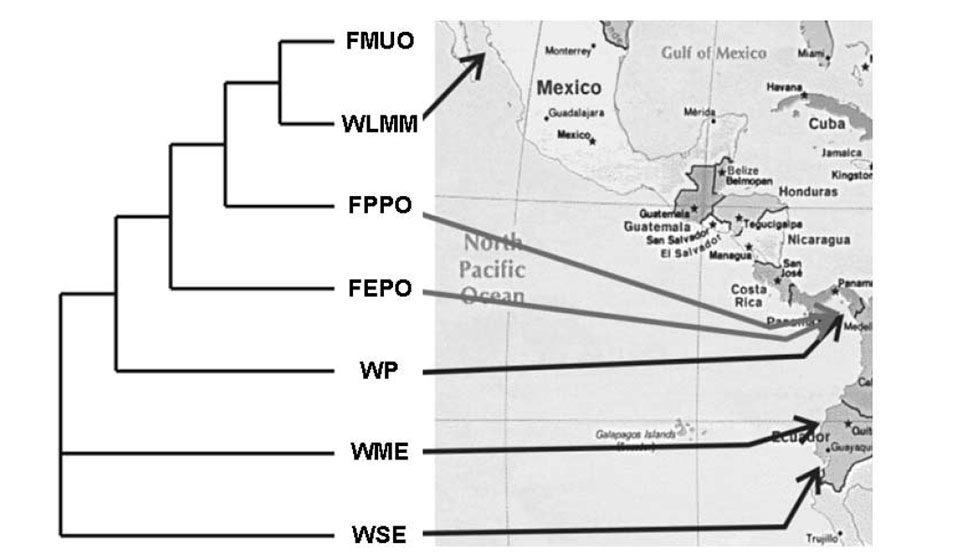
When comparing how captive stocks related genetically to the wild stocks of origin, the association was correct in every pairing, as displayed in Fig. 1. Finally, an assignment test was performed on the sample groups to determine how closely the populations were related (Table 2).
In all cases, the assignment of sample populations to a reference population based on genotype produced assignments exceeding 50 percent, and above 90 percent in two of the three farmed populations selected. The Mexican farmed sample of unknown origin were still assigned at 85 percent to the unknown category, but 13 percent related more closely to genetic profiles found in wild stocks found in Los Mochis, which is located less than 150 km from the source of the farmed animal samples.
Animals from the same founding wild stocks under different captive maintenance programs showed finite differences for identification purposes. Such differences can be used for “fingerprinting” pedigree and tracked irrespective of where they are farmed.
(Editor’s Note: This article was originally published in the February 2005 print edition of the Global Aquaculture Advocate.)
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-
Ken Jones, Ph.D.
Aquatic Stock Improvement Co. LLC
P.O. Box 5
Hawthorne, California 90250 USA[109,111,99,46,115,114,101,107,114,97,109,99,105,116,97,117,113,97,111,99,105,115,97,64,111,99,105,115,97]
-
Leland Lai
Aquatic Stock Improvement Co. LLC
P.O. Box 5
Hawthorne, California 90250 USA
Tagged With
Related Posts

Health & Welfare
DNA fingerprinting: System for black tiger shrimp
Molecular markers can allow on-farm selection of high-performance families without the need to use costly alternatives such as separate rearing of families/ groups of families or tagging animals.

Responsibility
A helping hand to lend: UK aquaculture seeks to broaden its horizons
Aquaculture is an essential contributor to the world food security challenge, and every stakeholder has a role to play in the sector’s evolution, delegates were told at the recent Aquaculture’s Global Outlook: Embracing Internationality seminar in Edinburgh, Scotland.

Health & Welfare
A look at aquaculture in Guyana
With its large quantities of water and little industry to pollute it, Guyana has the potential to become a greater player in global aquaculture.

Health & Welfare
‘Big picture’ connects shrimp disease, inbreeding
Disease problems on shrimp farms may be partly driven by an interaction between management practices that cause inbreeding in small hatcheries and the amplification by inbreeding of susceptibility to disease and environmental stresses.


