Newly described method an additional diagnostic tool for confirmation of the bacterial pathogen necrotising hepatopancreatitis

Necrotising hepatopancreatitis (NHP) is an enteric bacterial disease caused by a Gram-negative bacterium classified as Hepatobacter penaei (NHPB). It affects cultured penaeid shrimp in several countries from the Americas, including the United States, Mexico, Belize, El Salvador, Guatemala, Honduras, Costa Rica, Nicaragua, Panama, Brazil, Colombia, Ecuador, Peru and Venezuela.
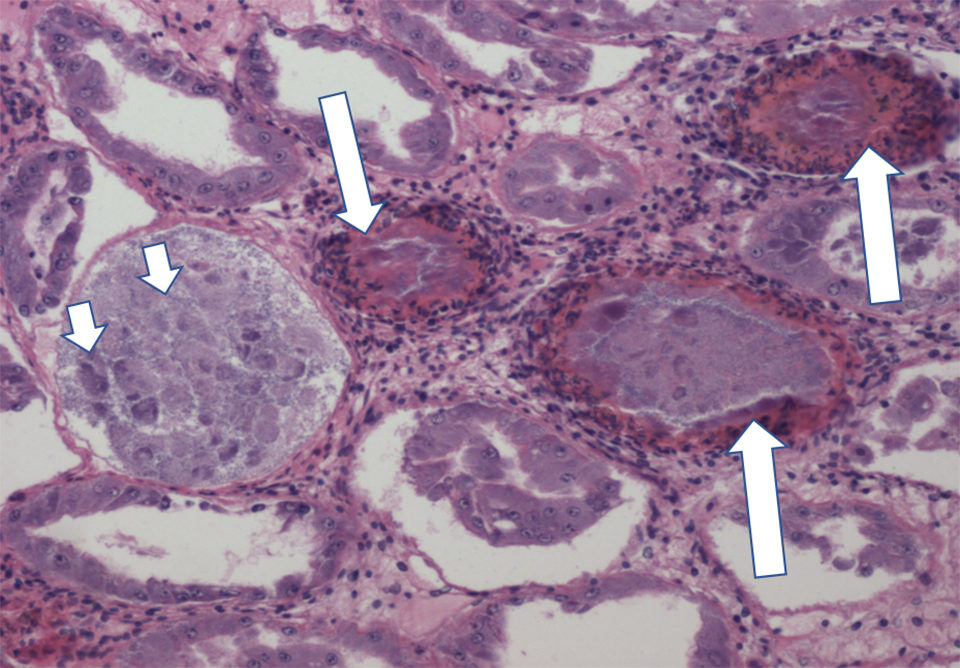
NHP is a chronic disease that causes mortalities up to 50 to 95 percent in affected postlarvae (PL), juveniles and broodstock of Pacific white shrimp (Penaeus vannamei). The manifestation of clinical signs in NHP at the farm level is dependent on environmental conditions such as high salinity and high temperature. NHP-infected shrimp show a typical soft shell, flaccid bodies, reduced feed intake and empty midguts.
The course of NHP infection has two phases: acute and chronic phases. Acute phase lesions in NHP-affected shrimp include necrosis and sloughing off of epithelial cells in the hepatopancreas (HP) and melanized HP tubules. In the chronic phase, the HP lesions are characterized by atrophy of tubules, reduced epithelial cell height, low lipid storage R cells and intratubular edema.
Since the first report in 1988, NHP has become such an important disease in the shrimp farming industry in the Western Hemisphere that, in 2010, it was listed in the List of Crustacean Diseases of the World Organization for Animal Health (OIE). Several diagnostic methods have been developed to detect and confirm the presence of NHPB, including Polymerase Chain Reaction (PCR), histology and in-situ hybridization and qPCR. However, there is only one PCR method recommended in the OIE manual and that creates the need to have an alternative PCR and real-time PCR (qPCR) assays for NHPB detection and confirmation.
The current PCR and qPCR assays based on the amplification of the 16S rRNA gene developed at The University of Arizona’s Aquaculture Pathology Laboratory (UAZ-APL) are the only methods recommended in the OIE manual for NHPB detection. Although these techniques are quite sensitive and specific to NHPB detection in shrimp, in recent years, occasionally non-specific amplifications have been observed in the end-point PCR when screening samples for NHPB in artemia cysts that were submitted to UAZ-APL.
Enhancing specificity of NHPB detection
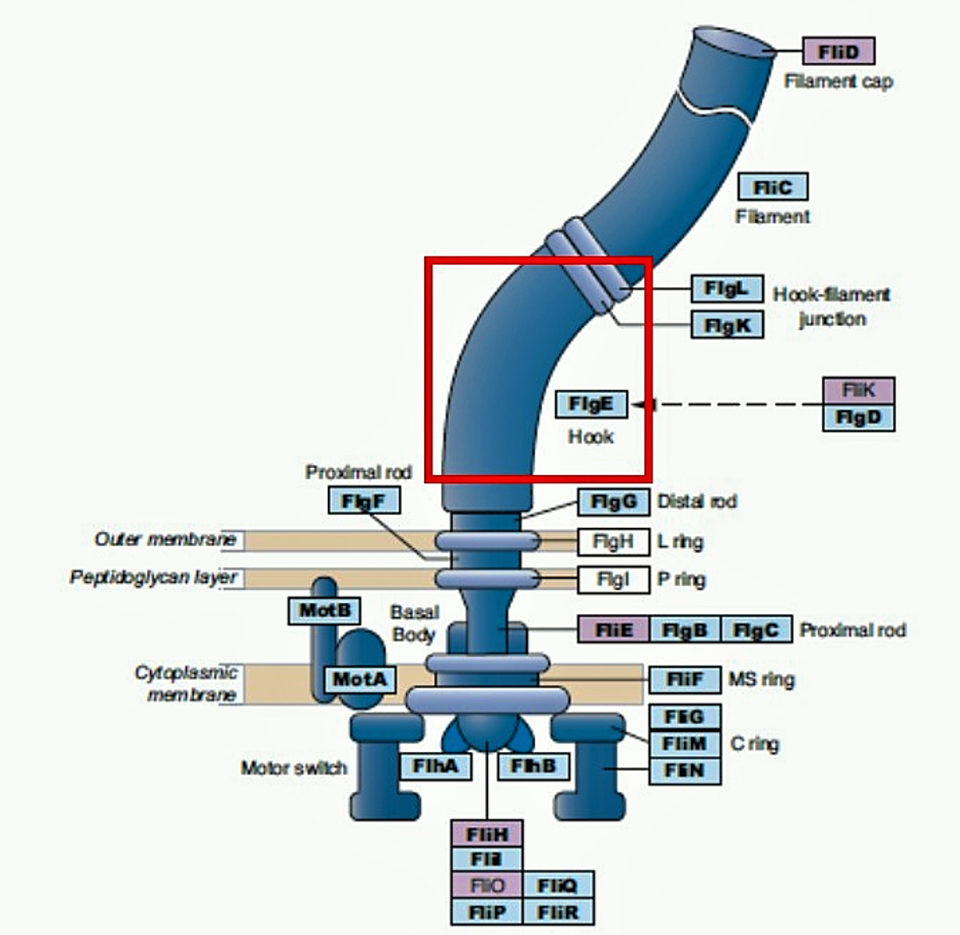
To enhance the specificity of NHPB detection by PCR and real-time PCR, we targeted a region of the NHPB flagella gene, called flagella hook protein (flgE), which is present only in flagellated bacteria. The flagella genes have evolved and diverged in a lineage-specific manner in the different bacterial groups, making this gene region highly specific for NHPB detection.
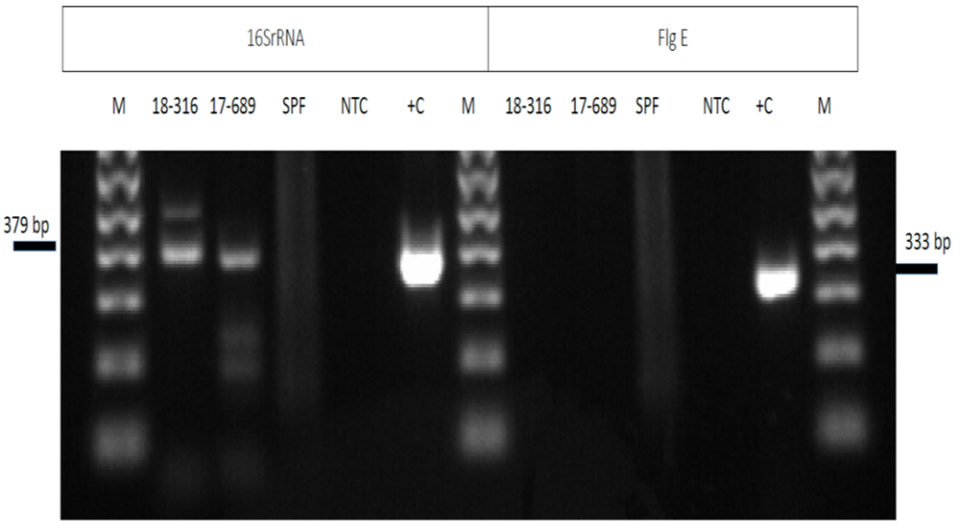
The agarose gel electrophoresis of PCR amplicons using the 16S rRNA primers from two samples (18-316 and 17-689 in the left panel of Fig. 3) of artemia cysts displayed some non-specific bands of 379 bp (Fig. 3). The panel on the right side shows the same set of samples amplified with the flgE primers. Unlike the amplification using the 16S rRNA primers, amplification with the flgE primers shows no amplification products in the electrophoresis gel. Sequencing of the amplicon from the 16Sr RNA primers confirmed the unspecific amplification product.
We also developed a new real-time PCR method targeting the same flgE region and by using a TaqMan hydrolysis probe (designed to increase the specificity of quantitative PCR). The new real-time PCR assay for NHPB did not provide any non-specific amplification, as obtained with the current OIE real-time PCR assay, and has a detection limit of 100 copies and a log-linear range of up to 108 copies.
Sensitivity
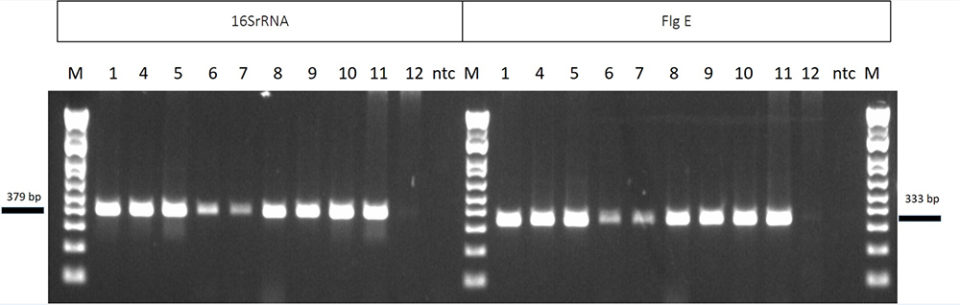
The analytical sensitivity of the new NHP PCR was determined by using 10 NHPB positive samples from seven origins with two different set of primers: 16S rRNA and flg E. In Fig. 4, the left side of the agarose gel shows samples amplified by using the current OIE-recommended method (16S rRNA); on the right side are the same samples amplified by the new method. Both methods provided similar results, and similar intensity of the DNA bands in the agarose gel.
In order to determine the specificity of amplification using the flgE gene-based protocol, DNA extracted from shrimp infected with viral diseases including IMN, YHD, IHHN, WSD and TS, and bacterial diseases – including V. parahaemolyticus, V. harveyi, Spiroplasma penaei – V.parahaemolyticus causing AHPND and NHPB were used as templates for PCR amplification. Negative results were obtained for all shrimp diseases except NHPB. This indicates the high specificity of the primers used for the amplification of flgE gene in NHPB detection.
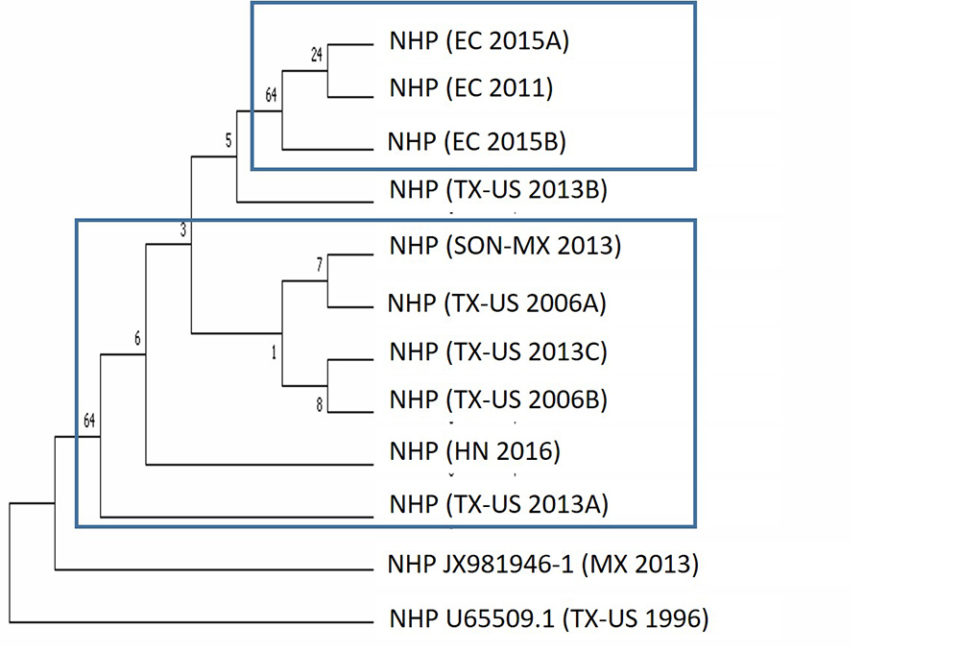
We also investigated the phylogenetic relationship of NHPB isolates obtained from different geographic regions. The NHPB-positive samples used in this study originated in Texas, USA (samples a & b from 2016), Ecuador (2011), Mexico (2013), Texas, USA (samples a, b & c from 2013), Ecuador (samples a & b from 2015) and Honduras (2016). The phylogenetic analysis based on the 16S rRNA gene showed two clusters: one cluster contained isolates from Ecuador, while the second cluster contained isolates from Mexico and Texas, USA (Fig. 5). Samples from Ecuador in 2011 and 2015 were grouped in the same cluster, which suggests the presence of the same NHPB isolate.
Implications of NHPB at the farm level
NHP has been reported in sub-adults and broodstock populations in farms, causing different mortality pattern in the Americas. In some countries, such as in Colombia, NHP does not cause acute mortalities as reported in other countries, and the typical NHP clinical signs are not always detected. This phenomenon could be related to the resistance of some P. vannamei lines to NHP. It is also possible that NHPB isolates may vary in their pathogenicity which could explain the different mortality pattern in different regions.
Perspectives
We described conventional PCR and real-time PCR assays based on the NHPB flgE gene as alternative methods for the detection and quantification of NHPB in shrimp and shrimp-associated samples, including artemia.
Both assays are highly specific and sensitive. The sensitivity of the flgE NHP PCR is similar to the currently recommended OIE method, but due to its higher specificity compared to the current OIE method, the newly described method will be an additional diagnostic tool for confirmation of this pathogen.
References available from corresponding author.
Authors
-

Luis Fernando Aranguren, Ph.D.
Corresponding author
Aquaculture Pathology Laboratory
School of Animal and Comparative Biomedical Sciences
The University of Arizona
Tucson, Arizona, USA -

Arun K. Dhar, Ph.D.
Associate Professor & Director
Aquaculture Pathology Laboratory
School of Animal and Comparative Biomedical Sciences
The University of Arizona
Tucson, Arizona, USA
Tagged With
Related Posts

Health & Welfare
Updates on shrimp diseases AHPND, NHP at Aquaculture America 2018
Several shrimp-disease papers were presented at Aquaculture America 2018, including one on the detection of AHPND at a site in the United States.

Health & Welfare
Wet mount technique excellent for health monitoring, not NHP diagnosis in shrimp
Necrotizing hepatopancreatitis is a disease of shrimp that causes slow growth, low survival and poor feed conversion. Wet mount analysis is commonly used to diagnose NHP in ponds in Brazil.

Health & Welfare
EHP a risk factor for other shrimp diseases
Laboratory challenges and a case-control study were used to determine the effects of EHP infection on two Vibrio diseases: acute hepatopancreatic necrosis disease (AHPND) and septic hepatopancreatic necrosis (SHPN).

Health & Welfare
The history and future of the Aquaculture Pathology Laboratory
The University of Arizona’s Aquaculture Pathology Laboratory has significantly contributed to the expansion of the shrimp farming for three decades.

