Results suggest potential benefits of microbial management of rearing water, feeds

The microbiota of animals is closely related to their health status, nutrient metabolism and immune system. After an animal hatches or is born, their intestinal, skin and oral microbial communities are gradually assembled. Microbial community assembly in early life may have a long-term impact on host health. And there is much interest in the relationship between the intestinal microbial community and the growth or health of aquatic invertebrates.
Larval production of the Pacific white shrimp (Litopenaeus vannamei) culture is impacted by various diseases that affect larval survival and quality, which is closely related to the growth, development and resistance to stress and/or disease of the animals.
Currently, research on the microbiome [all microbes, both helpful and potentially harmful ones, in an animal] of L. vannamei is mainly focused on other growth stages (i.e., juvenile, sub-adult, or adult) in terms of their associations with disease outbreak, growth and stress-resistance. Several studies have reported changes in the structure and function of the intestinal microbial community of postlarvae, juvenile and/or adult shrimp with development. An understanding on how the microbiota of L. vannamei functions during its early life is based on knowing the succession and assembly mechanism of larval microbial community.
Several previous, relevant studies with relatively low sampling frequency have yielded controversial results, indicating that an investigation at a fine temporal scale into larval shrimp microbiota in aquaculture practice is needed. Understanding the assembly mechanism of shrimp microbiota can help resolve the debate on whether we could improve the success rate of shrimp culture via manipulating their microbiota. However, little is known about the dynamics and taxonomic dependency of assembly processes of bacteria in larval shrimp.
This article – summarized and adapted from the original publication [Wang, Y., Wang, K., Huang, L. et al. Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome 8,106 (2020)] – discusses the results of a study using 16S rRNA gene amplicon sequencing [a technique used to reconstruct phylogenies (evolutionary relationships between species), due to the slow rates of evolution of this region of the gene] to investigate the succession and assembly processes of L. vannamei larval bacterial communities in a realistic aquaculture practice with sufficient biological replicates.
This work is funded by the Zhejiang Provincial Natural Science Foundation of China (LY18C030002), the National Natural Science Foundation of China (31672658), the Agricultural Major Project of Ningbo, China (2017C110001), the Basic Public Welfare Research Project of Zhejiang Province (LGN20C190008) and the K.C. Wong Magna Fund in Ningbo University.
Study setup
In this study, conducted in Wenchang, Hainan Province of China, L. vannamei were reared from eggs from the same pair of parents to minimize genetic divergence and inter-individual differences. The animals were reared in several 13-cubic-meter tanks in a larval rearing room under standard shrimp production conditions, including temperature 30 to 32 degrees-C; pH 8.0 to 8.3, salinity 30 to 33 PSU, and dissolved oxygen 5 to 8 mg per liter. Shrimp were stocked at 3.5 million nauplii in each tank.
Feeding of the developing larvae included commercial shrimp flakes at all stages after mouth opening (six times per day); at stages zoea I-II, live microalgae Chaetoceros sp. and Thalassiosira sp. were also used (three to four times per day). At the stages of zoea III and mysis, frozen brine shrimps (artemia) were used (three times per day), and then were replaced by live brine shrimp at the early postlarvae stage.
Both larvae and rearing water samples were collected across the nauplius, zoea I, zoea II, zoea III, mysis and early postlarvae stages lasting for 15 days (350 hours exactly). The developmental stages of larvae were confirmed by microscopy. Twenty-six sampling time points were used to collect both shrimp larvae and water samples, except at the 31st, 206th and 254th hour when only the larvae samples were collected. Sampled shrimp were processed for DNA extraction for analysis and other lab procedures.
For detailed information on the experimental design and sample collection; DNA extraction, 16S rRNA gene amplification, and other techniques used to analyze shrimp samples; and general ecological and statistical analyses, refer to the original publication.
Results and discussion
Our results showed that the larval bacterial communities originated mostly from the larval metacommunity [a set of interacting communities which are linked by the dispersal of multiple, potentially interacting species], while the water bacterioplankton community only contributed at the zoea sub-stages. These results suggest that the assembly of larval bacteria, overall, depends on the exchanges among individuals, probably via cross-feeding of feces and/or biofloc.
Shrimp larvae (especially at the zoea sub-stages) often have weak immune systems and are vulnerable to Zoea-II Syndrome, which can lead to high mortality. Some studies have shown that the diversity of animal intestinal microbiota is closely related to their functional integrity and stability. Although it has been suggested that the higher microbial diversity does not necessarily correspond to a more stable and healthier ecosystem, high diversity is often considered to help maintain the stability and ecological function of microbial community, thus being an important indicator of host health status. High bacterial diversity is desirable to maintain shrimp growth, health and stress-resistance.
The larval bacterial community composition varied with host development, and the high-frequency sampling strategy facilitated the unveiling of highly dynamic patterns of larval bacterial communities. Both the taxonomic and phylogenetic compositions of larval bacteria showed larval-stage-dependent patterns, even between the sub-stages of zoea. This is consistent with the pattern observed in the intestinal bacterial community of various fish larvae. In addition, we found distinct compositions and successional trajectories between larval and water bacterial communities. Similar results were previously reported for in juvenile and adult L. vannamei and the freshwater prawn Macrobrachium nipponense.
Our results showed that bacterial family Rhodobacteraceae had an overwhelming dominance over other bacteria groups after the mouth opening of the shrimp larvae. There were dynamic changes in bacterial community, represented by dramatic fluctuation in relative abundance of Rhodobacteraceae, Cyclobacteriaceae and Flavobacteriaceae (Fig. 1). It is likely that the differences in environmental conditions of diverse shrimp nursery systems may also lead to distinct patterns in larval microbiota dynamics. Future efforts are needed to reveal the impact of local environmental conditions on the succession of larval shrimp bacterial communities.
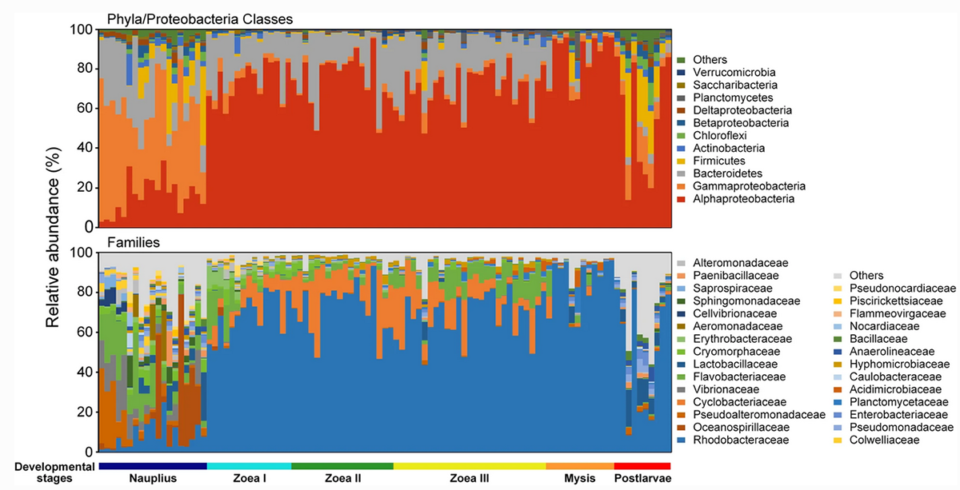
In our study, the shifts of physiological state, nutritional intake mode and microbial source between pre- and post-mouth opening stages of the shrimp larvae likely led to dramatic differences in larval bacterial communities. Results also suggest that the taxonomic turnover of bacteria was mainly driven by temporal switching among closely related taxa. Many studies have found Rhodobacteraceae persistently dominant in the intestinal tract of L. vannamei across juvenile to adult stages (ranging from 6 to 50 percent in average relative abundance). Therefore, the family Rhodobacteraceae is likely a core group of intestinal microbiota of L. vannamei.
Rhodobacteraceae are heterotrophic bacteria with extremely high diversity and versatility in organic matter degradation and widely distributed across various marine ecosystems. And most Rhodobacteraceae taxa are able to synthesize vitamin B12, which is a dietary essential for shrimp. Moreover, this family can significantly contribute to the potential in the biosynthesis and metabolism of multiple types of organic matter after the mouth opening of the larvae, participating in the metabolism of organic matters in the digestive tract of larvae and/or providing essential nutrients for host growth.
Also, the relative abundance of Rhodobacteraceae in the intestinal bacterial community of healthy L. vannamei individuals is often higher than that of diseased ones and shows an antagonistic relationship with potential pathogens such as Vibrio. And the Rhodobacteraceae might be a potential source of probiotics for larval shrimp nursery.
Collectively, the succession patterns and assembly mechanism of larval shrimp bacteria we determined in our study emphasize the importance of the shrimp mouth opening stage from the perspective of microbial ecology, indicating the possibility and timing of microbial management of the rearing water for larval microbiota regulation and pathogen prevention in commercial rearing systems of larval shrimp.
Perspectives
To the best of our knowledge, this study is the first systematic characterization on succession patterns and assembly processes of larval shrimp bacterial community at a fine temporal scale in an aquaculture setting.
Given the positive host selection for Rhodobacteraceae taxa from the rearing water during the zoea stage and its persistent dominance and large potential contribution to the metabolism of organic matters after the mouth opening of larvae, we suggest that Rhodobacteraceae could be crucial in the growth of shrimp larvae and thus be a potential source of probiotic candidates for larval nursery.
Based on the baseline of larval bacterial community succession we determined, future efforts should be made founded on a reduced sampling frequency and more comprehensive environmental profiles to better understand the impact of water quality and feeds on the succession and assembly of larval L. vannamei shrimp bacterial communities.
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Author
-
Dr. Kai Wang
State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products
Ningbo University
Ningbo, 315211, China; and
School of Marine Sciences
Ningbo University
Ningbo, 315211, China
Editor’s note: This article has nine co-authors (all listed at the end of the article); only the contact information of the corresponding co-author is included.
Tagged With
Related Posts

Health & Welfare
Characterizing bacterial communities in water, gut of Pacific blue shrimp
Study provides information about the bacterial community associated with biofloc and its influence on Pacific blue shrimp intestinal microbiota.

Responsibility
Bacterial amendments in shrimp grow-out ponds
Pond microbial communities are a critical and often overlooked component of aquaculture ecosystems. Bacterial amendments like probiotics provide significant support to shrimp farmers around the world.

Health & Welfare
Performance of Pacific white shrimp in biofloc with different light regimes
A study determined the zootechnical performance of Pacific white shrimp under different light regimes. Shrimp exposed to light had better zootechnical parameters than those with reduced or no exposure.

Aquafeeds
What is the best carbon-nitrogen ratio for biofloc systems?
A recent study at the Federal University of Rio Grande in Brazil showed that it is possible to reduce the C:N ratio in biofloc cultures, to decrease the water volume used and the total suspended solids produced, with resulting savings.


