Discipline incorporates diagnostic microarrays and analytical bioinformatics with principles of ecology

Oysters, one of the most heavily cultured aquatic organisms in the world, have historically been a viable economic resource based on the harvesting of natural populations. However, the health of two important species – the American oyster (Crassostrea virginica) and Pacific oyster (C. gigas) – is constantly under threat.
Parasitic infections result in diseases such as MSX (multinucleated sphere unknown) and dermo. Infectious diseases whose etiology remains somewhat of a mystery cause juvenile oyster disease or “summer mortality.” To face these challenges, researchers have developed a variety of cellular and molecular tools in an effort to gain the upper hand.
Polymerase chain reaction has greatly increased the ability to detect the presence of pathogens. Molecular techniques have been successful in isolating and characterizing the oyster genes involved in immune defense and disease resistance. These same techniques have also identified the virulence factors expressed by the pathogens, giving researchers targets for potential therapeutics. However, despite significant efforts, the mass mortality of native and cultured oysters remains one of the greatest threats to the industry.
Functional genomics
Within the last decade, major developments related to the field of molecular biology have culminated in an opportunity to expand understanding of the physiology of marine organisms at an accelerated rate. DNA microarrays, constructed by fixing a DNA template to a solid support such as a glass slide, are capable of holding copies of several thousand genes, allowing the simultaneous assessment of gene expression levels.
Microarrays can identify gene expression signatures – specific subsets of genes that respond to particular stimuli – that can be correlated with a variety of physiological responses. The strength of these correlations makes microarrays powerful diagnostic tools.
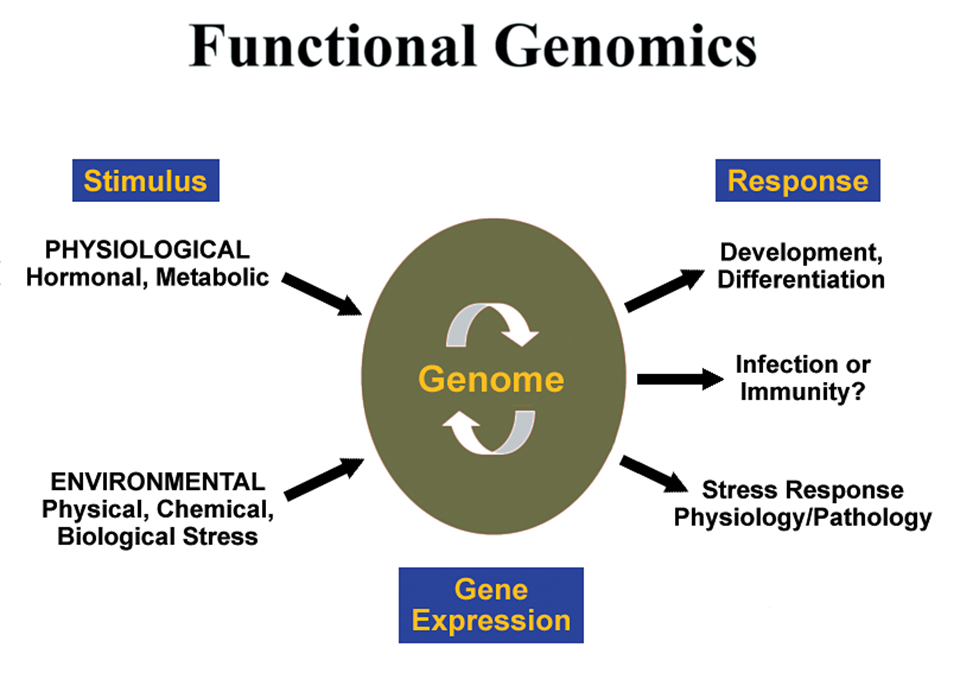
Complementary to this technology is the advancement of bioinformatics, a field grounded in statistics and empowered by computer technology, which enables the analysis of the large amounts of data generated by micro-arrays. These tools make a powerful combination for the basis of understanding the biological function of genes in “functional genomics.”
Microarrays: More than sexy new tools
Microarray technology offers unparalleled insight into the function of the entire genome. In most cases, technicians are interested in not only the response of individual genes, but also the dynamic interactions among genes. Analysis of such interactions is beyond the capacity of the linear models that have dominated quantitative genetics theory since its inception. Hence, the technological revolution produced by microarrays outstrips our ability to interpret the information with conventional statistics.
Developments in the analysis of complex systems, primarily artificial neural networks (ANNs) and the computation capacity of computer systems, offer new and powerful means to exploit the information content of microarray data. ANNs are universal modeling tools that are able to develop mathematical representations of virtually any data.
Artificial neural networks are capable of “learning” from data sets and providing predictive models of the performance of genotypes under any type of stress or set of environmental conditions. The strength of these models is in utilizing experience-based knowledge to make associations between environmental conditions, physiological parameters, performance characteristics, and gene expression profiles.
Valuable predictive models
The coupling of microarray technology with advanced computational tools is not as benign as it may seem. It requires a complete rethinking of experimental design and analysis, because ANNs are intended to provide predictive models under any arbitrary set of conditions. We do not want to repeat experiments as part of the model-building process, as the ANNs will be memorizing, not learning.
An even more heretical notion emerges from accepting nonrepetition. Does one want to use genetically related individuals to understand the impact of gene expression profiles on quantitative traits? The authors’ intuition says no. They have used related individuals in the past for this purpose because access to the genetic factors that produce the phenotype was largely denied.
Microarrays provide that access, and the use of unrelated individuals is more likely to provide a diverse set of responses upon which more general models can be constructed. A breeder may be very happy with a solution in which a set of animals under a defined set of conditions produces an amount of oysters. However, the value of models that could predict the production from genetically unrelated strains under varying environmental conditions can hardly be underestimated. Their application will require a rethinking of methods and analytical tools in a host of scientific disciplines.
Ecogenomics approach to oyster health
A vital component of estuarine systems, oysters play a role in determining water quality, productivity and plankton composition. All of these factors feed back into a natural loop to influence the health of oyster populations. Related to this loop is the common link found among all of the diseases that threaten oysters, which is the role of environmental factors in ultimately determining differences among survival, morbidity, and mortality.
Ecogenomics is a burgeoning discipline that incorporates the diagnostic power of microarrays and the analytical capabilities of bioinformatics with the basic principles of ecology to further understand the dynamic relationship between an organism and its environment. To achieve this goal, researchers from around the world interested in oyster genetics and genomics have come together to develop a microarray comprised of genes from both C. virginica and C. gigas, as well as genes and specific markers from known pathogens responsible for common oyster diseases.
Furthermore, the inclusion of the C. gigas genes further elucidates the mechanisms involved in pathogen interaction, since this particular species of oyster appears to be more resistant to diseases that threaten C. virginica. The use of a multispecies microarray offers researchers the opportunity to assess host-pathogen interactions at the molecular level.
Conclusion
If we consider the advances made in agriculture, it is evident that genomics research will play an increasingly important role in the future of molluscan aquaculture. The use of microarrays opens up a vast panorama of possibilities to researchers and aquaculturists.
Microarrays can assess differences in quantitative traits between strains, elucidate mechanisms of disease resistance, accelerate selective-breeding regimes, assess the impacts of environmental conditions, and establish early indicators of disease that are capable of accurately predicting the impact on survival.
(Editor’s Note: This article was originally published in the June 2004 print edition of the Global Aquaculture Advocate.)
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-
Matthew J. Jenny
Marine Biomedicine & Environmental Sciences Center
Medical University of South Carolina
Hollings Marine Lab
331 Fort Johnson Road
Charleston, South Carolina 29412 USA -
Robert W. Chapman, Ph.D.
South Carolina Department of Natural Resources
Marine Resources Research Institute
Charleston, South Carolina, USA
Tagged With
Related Posts

Intelligence
Aquaculture has put the oysters back in Oyster Town
Whitstable Oyster Fishery Co. aims to safeguard the English town’s rich oyster tradition. Farming triploid oysters on racks in intertidal zones has made a "massive difference," says the company's managing director.

Intelligence
Behold the nutritious oyster
Oysters provide important, natural filtration of water and are an important component of many healthy coastal ecosystems because their active filtering can help improve and maintain water quality. For many coastal communities, oysters are an important food resource and excellent sources of protein and amino acids, zinc, selenium, iron and B-vitamins.

Responsibility
Ailing waterways hail the oyster’s return
The Lower Hudson Estuary and Chesapeake Bay, two waterways once home to thriving oyster beds, would welcome the shellfish’s return. Aquaculture initiatives in both areas aim to reinvigorate the water and the communities they support.

Responsibility
Can oyster farms protect wild oysters from disease?
Research results suggest that oyster farming can enhance wild oyster populations if oysters are harvested before spreading disease.


