Results show influence of abiotic, biotic factors

Among the infectious diseases of penaeid shrimp, the white spot syndrome (WSS) and the acute hepatopancreatic necrosis disease (AHPND, previously referred as “early mortality syndrome” or EMS) are currently the most serious threats to shrimp farmers. WSS is caused by the white spot syndrome virus (WSSV), whereas the AHPND is an emerging bacteriosis caused by virulent strains of Vibrio parahaemolyticus and V. harveyi. The adoption of on-farm biosecurity practices is necessary to limit the pathogen entrance into the culture systems.
Among on-shrimp farm biosecurity practices, the biofloc super-intensive technology (BFT) is a promising alternative culture system. It is widely believed that BFT culture improves the crustacean immunity leading to high survival rates even under bacterial and viral infections. Although the mechanisms underlying shrimp robustness are not yet understood, a continuous immunostimulation condition is rather expected considering the abundance of microbial-associated molecular patterns (MAMPs) present in BFT systems which can activate innate immune responses.
Since the aquatic environment can influence the microbiota composition and abundance, studies focusing the BFT contribution on the establishment of shrimp intestinal microbiota are highly required. In addition, it is now well established that commensal microbiota is essential for the correct functionality of the host physiology. Surprisingly, the characterization of the microbial communities present in the digestive tract of cultured shrimp species has been only recently uncovered.
To our knowledge, only one report regarding the description of the intestinal bacterial communities of a penaeid species – the Pacific blue shrimp (Litopenaeus stylirostris) – reared in a BFT system is available in the literature. In addition, nothing is known about the bacteriome (a specialized organ with specialized cells that provide shelter and nutrients to the bacteria while protecting the host animal) plasticity in shrimp infected by the WSSV, one of the most important pathogens in shrimp farming.
This article – adapted and summarized from the original publication – was designed to characterize the abundance and composition of the intestinal bacterial communities of the most important penaeid species, the Pacific white shrimp (L. vannamei) reared in BFT and a clear seawater system. Likewise, the plasticity of the midgut bacteriome from shrimp challenged by WSSV was investigated. Our results bring new evidence of the influence of the biofloc culture and the viral challenge on the shrimp bacteriome, providing new insights into future studies regarding the role of microbiota on the intestinal immunity of cultured penaeid.
Study setup
L. vannamei postlarvae (five days old, PL5) from a commercial shrimp hatchery (Aquatec LTDA Canguaratema, Rio Grande do Norte, Brazil) were used in this study at the Laboratory of Marine Shrimp (Federal University of Santa Catarina, Florianópolis, Brazil). The biofloc culture was initially established in a 50-cubic-meter matrix tank and the experimental design is shown in Fig. 1.
Shrimp PLs were randomly stocked into eight, 9-cubic-meter indoor tanks, with four BFT tanks and four clear seawater system tanks, at an initial stocking density of 300 and 20 PL5 per cubic meter, respectively. The tanks were continuously aerated (dissolved oxygen > 5 mg/L) and kept under controlled temperature (29 ± 1 degrees-C) and salinity (34 to 35 ppt).
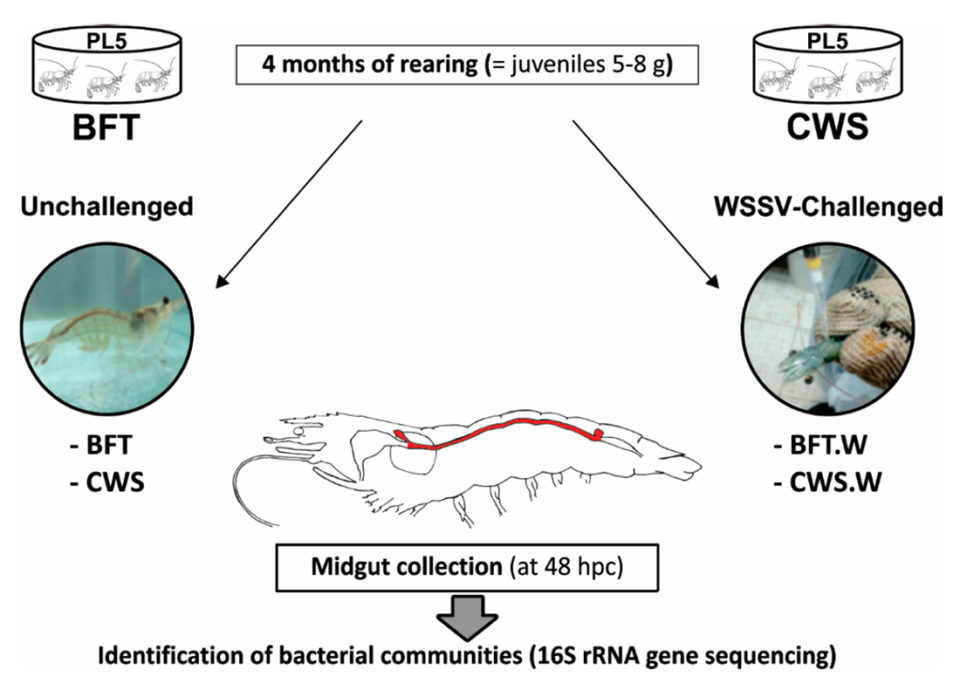
Post-larvae were fed four times a day with a commercial diet (Guabi Potimar), and tank water was exchanged at 80 percent daily. After four months, when the shrimp had become 5- to 8-gram juveniles, around 30 percent of the animals from each tank/group were randomly selected to confirm that the shrimp were free of WSSV by using the nested-PCR assay, and then 120 animals from each culture system were transferred to the Laboratory of Immunology Applied to Aquaculture (Federal University of Santa Catarina, Florianópolis, Brazil) for various tests and analyses.
For a detailed description of the experimental design; WSSV per os (oral) challenge and midgut collection; genomic DNA (gDNA) extraction; 16S rRNA gene library preparation and high throughput sequencing; and sequence data analysis, refer to the original publication.
Results and discussion
Our study explored the bacterial communities’ dynamics (abundance and phylogenetic composition) in the shrimp midgut in response to two important abiotic and biotic factors related to shrimp farming (culture system and viral infection) by assessing 16S rRNA gene sequencing [this technique is used in reconstructing phylogenies – evolutionary histories – due to the slow rates of evolution of this region of the gene]. We generated two bacterial 16S rRNA gene libraries from the midguts of shrimp cultured in a BioFloc Technology (library “BFT”) and in a clear seawater system (library “CWS”). Likewise, two other libraries were generated from the midguts of shrimp challenged with the WSSV using a per os method (libraries “BFT.W” and “CWS.W”).
Analysis of Venn diagrams [which show all possible logical relations between a finite collection of different sets] revealed significant differences in the frequency distribution of bacterial operational taxonomic units (OTU; operational definition used to classify groups of closely related individuals) according to the culture system (BFT and CWS) and viral challenge (Fig. 2). The midguts of animals reared in BFT exhibited a larger number of OTUs when compared with those from CWS. Five-hundred and seventy-one OTUs were exclusively found in shrimp reared in bioflocs (361 in BFT, 111 in BFT.W and 99 in both groups), whereas 298 OTUs were exclusive from shrimp reared in clear seawater (162 in CWS, 121 in CWS.W and 15 in both conditions).
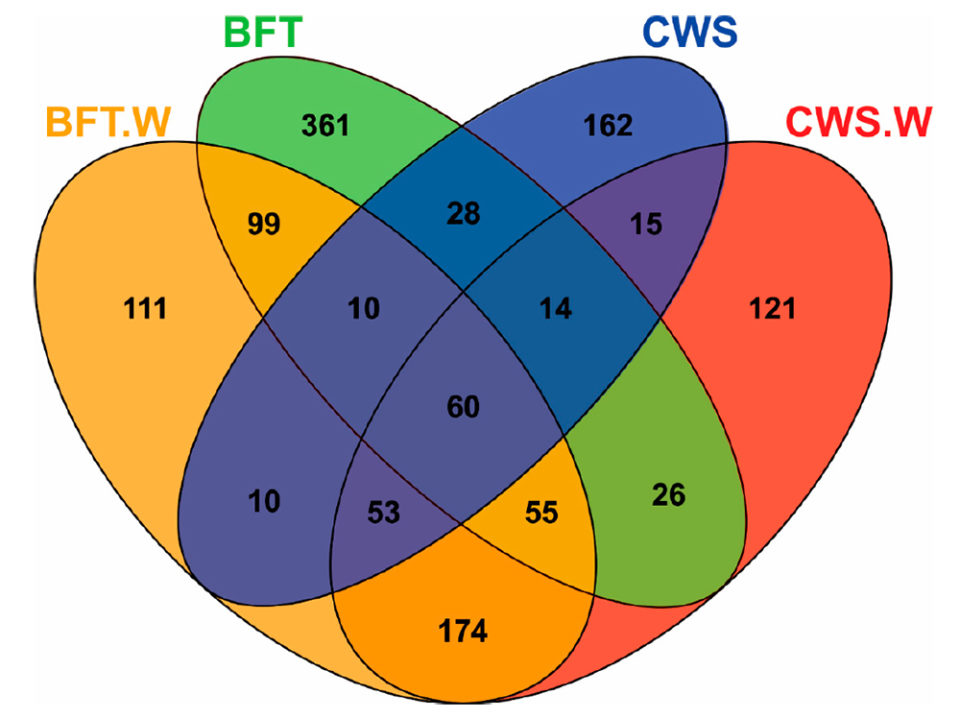
The higher number of exclusive OTUs from the BFT samples could reflect the diversity microbioma of the biofloc environment. The viral challenge led to the appearance of exclusive OTUs in each rearing condition: 111 OTUs for BFT.W and 121 for CWS.W. Furthermore, exclusive OTUs (n = 174) were shared only by the challenged animals of both rearing conditions. These findings suggest that this bacterial community displacement in the midgut is related to the virus presence. Finally, 60 OTUs were shared among all samples, which represented 4.61 percent of the total OTUs. This bacteria subset present in all groups could represent relevant microorganisms to the fundamental structure and function of the shrimp intestinal microbiota. Additional studies on microbiota shrimp-virus interaction related to the BFT rearing system deserve to be investigated in the future.
Regarding the influence of rearing conditions on the bacterial communities of shrimp midgut, it is widely believed that the commensal microbiota composition in adult arthropods [invertebrate animals with an exoskeleton (external skeleton), like insects and crustaceans] appears to be intimately related to the initial exposure to microorganisms during their early life. We therefore cultured our shrimp in BFT and CWS for four months (PL5 to 5 to 8 grams) before the lab testing. In our analysis, the resulting high-quality 16S rRNA gene sequences were classified into 33 prokaryotic phyla that belong to the domain Bacteria. The most representative phyla identified in the L. vannamei midgut were Proteobacteria, Bacteroidetes, Actinobacteria and Firmicutes. However, the frequency distribution of the intestinal bacterial communities differed according to the rearing system (Fig. 3).
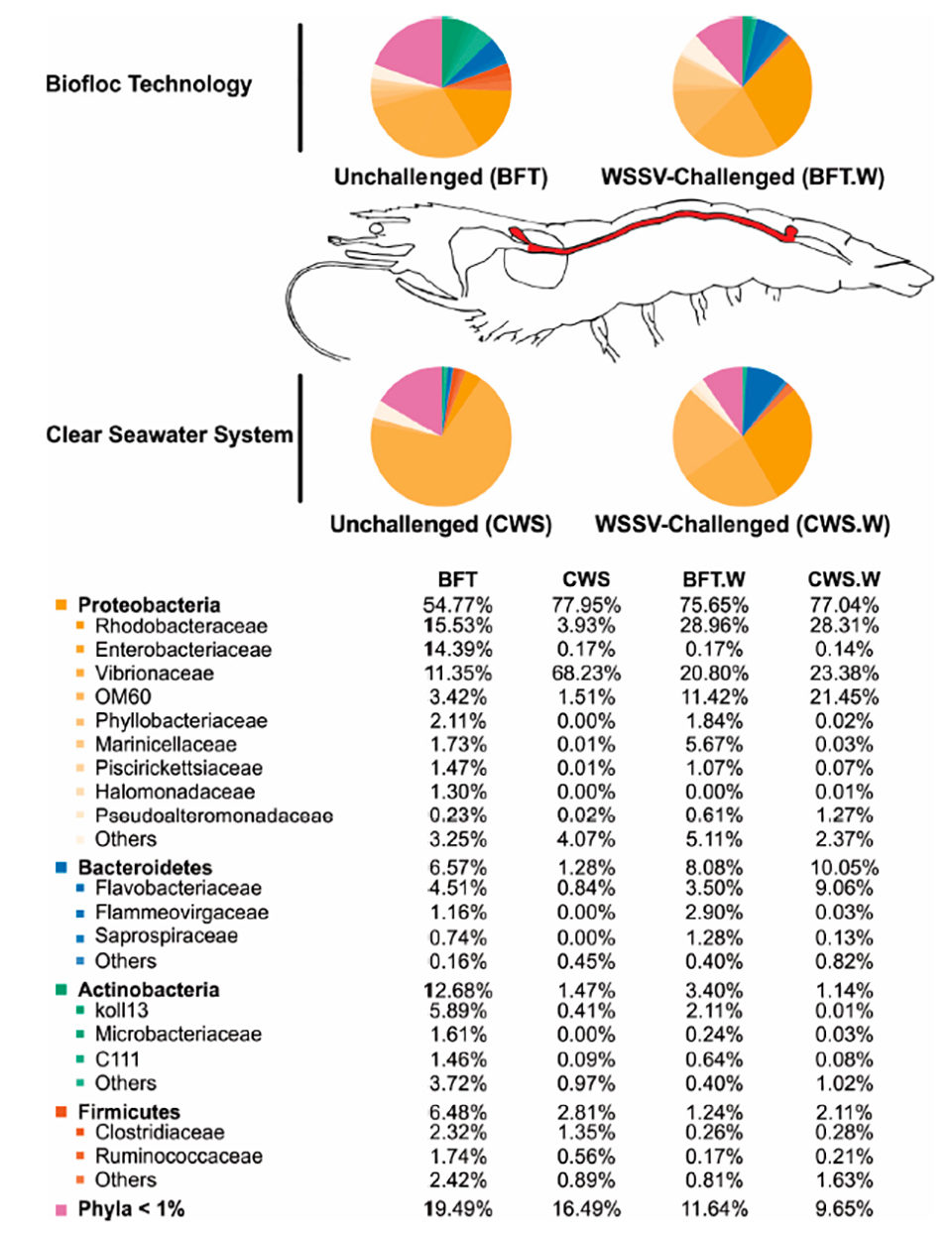
The genus Vibrio is composed of fast-growing aquatic bacteria able to colonize the digestive tract of different animals, including penaeids. Many Vibrio species are opportunistic pathogens in shrimp under stressful conditions, such as poor nutrition, low water quality, and immune depression. In cultured shrimp, this bacterial group has been repeatedly implicated in gastro-intestinal diseases, leading to high shrimp mortalities. Microorganisms present in BFT water could act against pathogenic bacteria by competition for substrate and nutrients, producing inhibitory compounds, and interfering in the bacterial quorum-sensing [a system of stimuli and response correlated to population density, used by many bacteria to coordinate gene expression according to the density of their local population] communication.
Interestingly, two Proteobacteria families, Rhodobacteraceae and Enterobacteriaceae, were more abundant in the midgut of shrimp reared in BFT than in CWS, representing around 15 percent in BFT and less than 4 percent in CWS. The role of this bacterial family in the shrimp intestinal microbiota is not well understood, but it is believed that the BFT system can favor the presence of this bacteria family due to its high concentration of suspended solid that can be used as growth sites by Rhodobacteraceae. Family members can establish an antagonistic activity limiting the survival of pathogenic Vibrio. Therefore, we could hypothesize that the higher abundance of Rhodobacteraceae in the midgut of L. vannamei reared in BFT could be associated with a lower abundance of Vibrionaceae.
Our findings indicate that the bacteriome of shrimp reared in BFT was more diversified and richer when compared to that from animals reared in clear seawater, where the predominant bacterial community was Vibrionaceae. The water from BFT is especially rich in organic matter and suspended particles, which can favor bacteria that use organic matter and nitrogen compounds for growth. In addition, the BFT rearing system apparently causes important modifications in shrimp midgut microbiota compared to CWS, which corroborates the fact that the microbiota from the digestive tract of the aquatic animals is directly influenced by the environment. In addition, the idea of considering the BFT as a “natural probiotic system” has important consequences to the intestinal microbiota. The BFT could act internally and/or externally to the shrimp body, an effect promoted by large groups of microorganisms, but mainly bacteria.
Regarding the beneficial effects of the BFT system, bioflocs can act as immunostimulants and enhancing the shrimp innate immune system, even altering the expression of genes related to the shrimp immune response, which could be attributed to the ability of the BFT to induce changes in shrimp microbiota. Overall, our results show that the shrimp gut microbiota is altered by the rearing environment.
Regarding the shrimp intestinal microbiota plasticity in response to a viral challenge, the most impacting results of our study were to characterize the bacterial communities shift in the shrimp midgut challenged by WSSV. To the best of our knowledge, there is only one study on the shift of gut microbiota in response to the WSSV infection, evaluated in the Chinese mitten crab. Ours is the first study investigating the effect of a viral pathogen on the intestinal microbiota of a penaeid species reared in a BFT system.
Perspectives
By using a high-throughput sequencing technology, we have characterized the intestinal bacteriome of the most important cultivated shrimp species, L. vannamei, and assessed the influence of the BFT rearing and of the WSSV challenge on the composition and abundance of the bacterial communities.
The bacterial composition from the midgut of shrimp reared in bioflocs was richer and more diverse than that from clear seawater. The predominant bacterial group belonged to the phylum Proteobacteria (Rhodobacteraceae, Enterobacteriaceae and Vibrionacea), followed by the phyla Bacteroidetes (Flavobacteriaceae), Actinobacteria and Firmicutes. Vibrionaceae was more abundant in the CWS group than in BFT-reared shrimp (68.23 percent and 11.35 percent from total bacterial communities, respectively).
The bacterial composition of L. vannamei midgut was affected by the WSSV challenge. Vibrionaceae was the most affected bacterial family and its abundance doubled in the midgut of BFT-reared shrimp after viral challenge, while in CWS-reared shrimp decreased drastically. In addition, the WSSV challenge apparently led to a more homogeneous distribution of bacterial population composition, as Rhodobacteraceae, Enterobacteraceae and Vibrionaceae, in the midgut of shrimp reared in both culture systems, BFT and CWS. The changes in the gut bacteria diversity associated to the WSSV challenge could indicate a displacement in the intestinal microbial communities leading to the dysbiosis (a microbial imbalance or maladaptation on or inside the body, like an impaired microbiota) condition.
Knowing the intestinal bacterial populations of shrimp reared in BFT and during WSSV infection is a relevant step to understanding the role of intestinal bacteriome microbiota on crustacean immune defenses against viral diseases. Although the molecular mechanisms involved in the control and regulation of the shrimp gut microbiota is still largely unknown, the environmental conditions and the presence of infectious agents proved to be decisive factors influencing both the diversity and abundance of the bacterial communities.
With these 16S rRNA sequencing data in hand, and given that penaeid shrimp is an excellent model for functional genomic studies, we can now investigate shrimp in host-microbiota interactions, and also the role of the commensal microbiota in the regulation of the shrimp gut immunity.
Now that you've finished reading the article ...
… we hope you’ll consider supporting our mission to document the evolution of the global aquaculture industry and share our vast network of contributors’ expansive knowledge every week.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year. GSA individual and corporate members receive complimentary access to a series of GOAL virtual events beginning in April. Join now.
Not a GSA member? Join us.
Authors
-
Dr. Mariana R. Pilotto
Laboratory of Immunology Applied to Aquaculture
Department of Cell Biology, Embryology and Genetics
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil -
Dr. André N. A. Goncalves
Laboratory of Immunology Applied to Aquaculture
Department of Cell Biology, Embryology and Genetics
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil -
Dr. Felipe N. Vieira
Laboratory of Marine Shrimp, Department of Aquaculture
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil -
Dr. Walter Q. Seifert
Laboratory of Marine Shrimp, Department of Aquaculture
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil -
Dr. Evelyne Bachère
Ifremer, UMR 5244, IHPE Interactions-Hosts-Pathogens-Environment, UPVD, CNRS
Université de Montpellier, 34095 Montpellier, France -
Dr. Rafael D. Rosa
Laboratory of Immunology Applied to Aquaculture
Department of Cell Biology, Embryology and Genetics
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil -
Dr. Luciane M. Perazzolo
Corresponding author
Laboratory of Immunology Applied to Aquaculture
Department of Cell Biology, Embryology and Genetics
Federal University of Santa Catarina
88040-900 Florianópolis, SC, Brazil
Tagged With
Related Posts

Health & Welfare
The myth of the static microbiome
In these early stages of understanding how microbiome changes impact animal health, use caution in interpreting lab results and how they may apply to the real world.

Responsibility
Aquaculture byproducts improve sustainability of seafood value chains
Tons of aquaculture byproducts are available as sources for fishmeal and fish oil to supplement the supplies obtained from fisheries. Innovative technologies are supporting more efficient use of these by-products in aquafeed.

Health & Welfare
Big shoes to fill: Dhar takes reins at shrimp pathology laboratory
Arun Dhar, Ph.D. will attempt to fill the “big shoes” of Dr. Donald Lightner at the University of Arizona’s Aquaculture Pathology Laboratory, where the shrimp disease EMS was diagnosed.

Health & Welfare
A holistic management approach to EMS
Early Mortality Syndrome has devastated farmed shrimp in Asia and Latin America. With better understanding of the pathogen and the development and improvement of novel strategies, shrimp farmers are now able to better manage the disease.


