University of Arizona conducts robust disease diagnoses for producers

Based on the importance of infectious diseases in the shrimp industry worldwide and the problems associated with their presence, the diagnosis of shrimp pathogens has become one of the most critical components for this growing industry. A positive result from a particular diagnostic test for any of the diseases listed by the World Organization for Animal Health (OIE). can have a dramatic consequence for shrimp farming at different levels:
At a national level
For OIE-member countries, there is a requirement to report to this organization. Even though some countries do not report promptly to the OIE, when it happens it could lead to some trade restrictions or additional requirements before exporting shrimp or manufactured shrimp products from the corresponding country.
At a quarantine level
Usually the quarantine process is being carried out when live shrimp is imported. As a part of the process, sampling of dead/symptomatic and live shrimp is conducted in the importing country to confirm the sanitary status. When a given sample tests positive, restrictions are imposed on the corresponding exporting supplier/country.
At broodstock/hatchery level
Like any other animal-producing industry, shrimp broodstock are the backbone of the shrimp industry. When a given broodstock/postlarvae sample tests positive for any of the OIE-listed diseases, the consequences could be as extreme as the elimination of the entire broodstock/postlarvae stock.
At grow-out pond level
Depending on the local and regional biosecurity strategies, the consequences of a positive result could lead to either emergency harvest or elimination of the population.
Disease diagnosis as a part of the biosecurity components
One of the main components of biosecurity in the shrimp industry is disease diagnosis analysis as a mean to validate biosecurity status, and includes:
- Knowledge of diseases of concern.
- Determining the list of diseases/pathogens that need to be excluded.
- Adequate diagnostic/detection methods.
- Use of “clean” shrimp stocks.
- Assurance/surveillance of cultured stocks.
This implies that having a reliable diagnostic lab for shrimp pathogen detection is an integral part of the biosecurity strategy for the shrimp industry. According to the OIE manual, the preferred diagnostic method for shrimp pathogens detection is the polymerase chain reaction (PCR) test, a molecular biology technique to amplify a single or a few copies of a segment of DNA across several orders of magnitude to generate many (thousands to millions of) copies of a particular DNA sequence. The PCR technique is preferred for several reasons including: availability, utility and diagnostic specificity, and sensitivity. In Table 1 the PCR information for each shrimp pathogen is grouped and compiled from the OIE manual 2017.
As shown in Table 1, PCR is the preferred method for all shrimp pathogens detection for presumptive and confirmative diagnosis in all the stages. The only exception is the WSSV detection by PCR at the larval stage, for which there is not an OIE-recommended method.
Aranguren, PCR detection, Table 1
| Pathogen | Larvae | PLs | Juvenile | Adult | Presumptive diagnosis | Confirmatory diagnosis |
|---|
Pathogen | Larvae | PLs | Juvenile | Adult | Presumptive diagnosis | Confirmatory diagnosis |
|---|---|---|---|---|---|---|
| IHHNV | a | a | a | a | a | a |
| WSSV | d | b | a | a | a | a |
| MBV | a | a | a | a | a | a |
| BP | a | a | a | a | a | a |
| YHV | a | a | a | a | a | a |
| TSV | a | a | a | a | a | a |
| IMNV | a | a | a | a | a | a |
| AHPND | a | a | a | a | a | a |
| NHP | a | a | a | a | a | a |
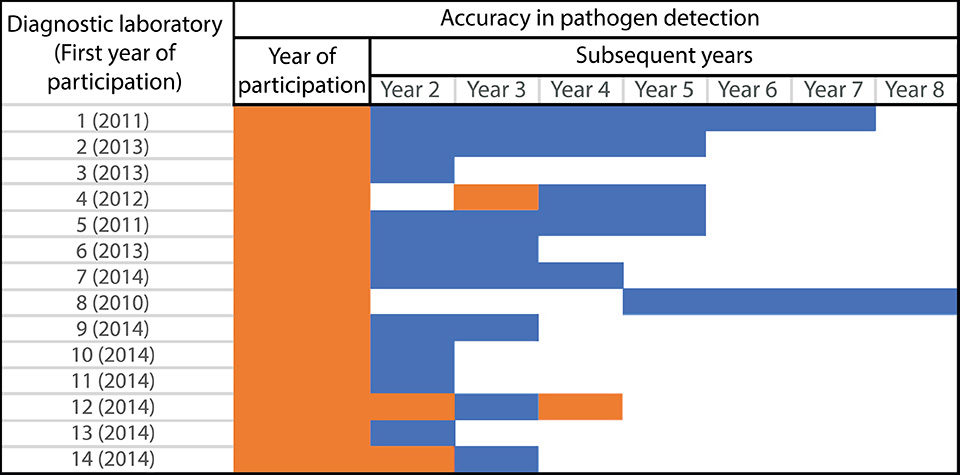
Proficiency Test or Ring Test (PT/RT)

Since 2005, The University of Arizona Aquaculture Pathology Laboratory (UAZ-APL; Arizona USA) has carried out the Proficiency Test (PT), also called the Ring Test (RT). Since then, around 301 diagnostic laboratories have participated in the PT, which has helped them improve their capabilities in the diagnosis of shrimp pathogens by PCR.
Based on the data collected over the years, many times when a laboratory participates in a PT for the first time, the corresponding diagnostic laboratory usually presents some issues with false positive (+) or false negative (-) results (Fig. 1). However, as the same laboratories continue to participate in the PT, they tend to improve their diagnostic accuracy. Fig. 1 shows a comparison of initial participation of some laboratories in the PT and their progress in the subsequent participations by the same laboratories.
It is abundantly clear from the data collected over the years that the PT can serve as an indicator that can help improve the disease diagnosis accuracy of a diagnostic laboratory. The UAZ-APL conducts PTs twice per year with the following features:
- Participation is voluntary.
- Results are confidential.
- There is no prescribed method for a particular disease detection. Therefore, the participating laboratories select their own method of detection.
- Periodicity: Twice a year (February and August).
- Sample shipping must comply with importation requirements specific to each country.
- A panel of 10 coded samples is provided, which contains non-infectious tissue of the pathogens listed in Table 2.
- The participating laboratory may choose to carry out testing from one to all the pathogens.
- A standardized format to report the results. The results must include primer sets/commercial kits, extraction methods, PCR conditions, gel photographs, amplification plots and melting curves, among others.
- Time to return results: Seven working days from the time moment samples are received by a participating laboratory.
- The University of Arizona reports the results to each participant, and recommendations are provided to further improve disease detection accuracy, if needed.
Aranguren, PCR detection, Table 2
| Pathogen | OIE | Category |
|---|
Pathogen | OIE | Category |
|---|---|---|
| White spot syndrome virus (WSSV) | Yes | C1 |
| Taura syndrome virus | Yes | C1 |
| Yellow head virus genotype 1 (YHV1) | Yes | C1 |
| Infectious hypodermal and hematopoietic necrosis virus (IHHNV) | Yes | C3 |
| Infectious myonecrosis virus (IMNV) | Yes | C1, C2 |
| Necrotizing hepatopancreatitis (NHPB) | Yes | C2 |
| Penaeus vannamei nodavirus (PvNV)* | No | C2, C3 |
| Vibrio parahaemolyticus causing Acute hepatopancreatic necrosis disease AHPND /EMS (VPAHPND) | Yes | C1 |
| Enterocytozoon hepatopenaei (EHP) | No | C2 |
| Specific pathogens free (SPF) | NA | NA |
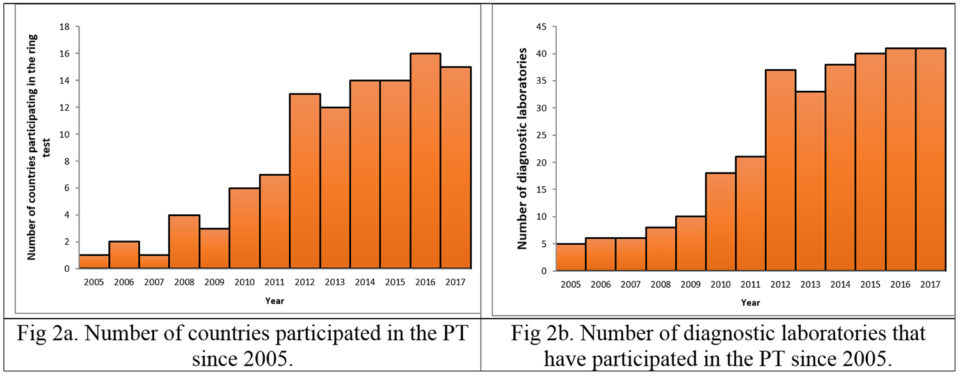
Since the year 2005, several diagnostic laboratories from all over the world have participated in the PT. Fig. 2 (panels a and b) shows disease diagnostic laboratory from different countries that have participated in the proficiency test conducted by UAZ-APL since 2005. In 2017, 41 diagnostic laboratories from 15 countries participated in the PT.
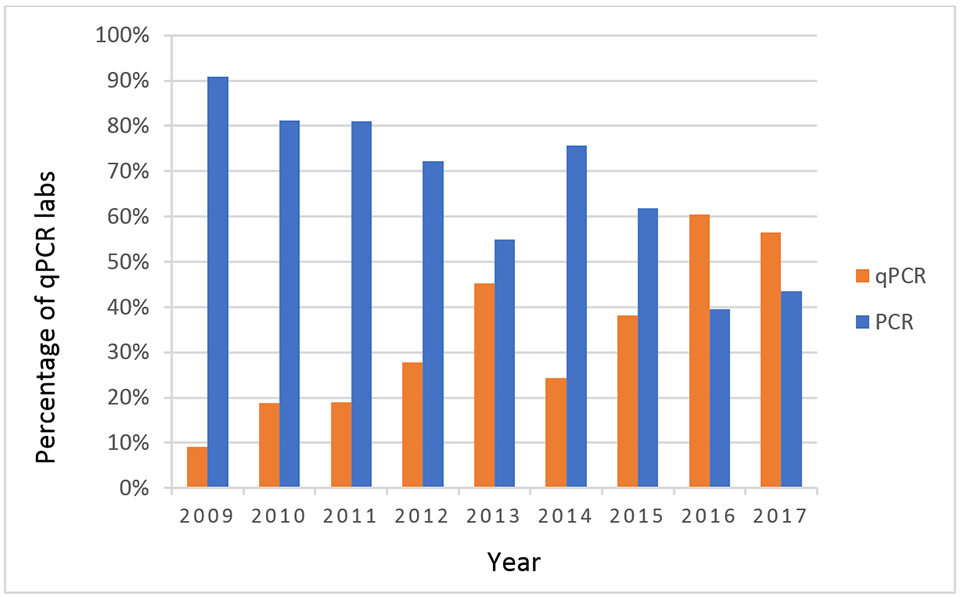
The data collected for the PT over the years show a developing trend in shrimp pathogen detection methods. Fig. 3 shows a change in the trajectory of pathogen detection by PCR from end point PCR to real-time PCR (qPCR). The data collected show that, in 2009, less than 10 percent of the laboratories were using qPCR for pathogen detection, and in 2017 more than 50 percent of the diagnostic laboratories have adopted qPCR as their standard method for pathogen detection.
Common problems encountered in shrimp pathogen detection as reported in Proficiency Test results
A review and subsequent analyses of the results reported by participating laboratories over the years revealed some common problems associated with the PT conducted by the participating laboratories. Listed below are some common problems that we have identified in the results reported:
Sample switching
Sometimes a participating laboratory carries out sample processing correctly. However, at the time of result submission, the positive samples are reported with a different code. In addition, it appears that during the first step of the sample processing, i.e. nucleic acid isolation, sample switching had occurred. As a result, original samples are now mislabeled, and obviously a mislabeling of the samples can cause an error in result reporting.
False negative

This is caused by several reasons, including:
- Reverse transcription step is not working. While detecting RNA pathogens by PCR, this is a common problem. In order to make a cDNA from an RNA template, the reverse transcriptase enzyme is used. Reverse transcriptase is a thermolabile enzyme. Due to an improper storage, this enzyme does not work efficiently or simply does not work at all. If this does not happen, the RNA genome of the pathogen will not be reverse- transcribed and the PCR amplification will not yield any product, resulting in a false negative.
- The IMNV virus, a dsRNA pathogen. For this particular virus, an additional step is required before starting cDNA synthesis. Boiling the sample in order to denature the RNA is a pre-requisite for cDNA synthesis. If the RNA sample is not boiled, false negative results will occur.
- High concentration of DNA. When a high concentration is present in the PCR, it could inhibit the amplification reaction. It is relatively easy to note the high concentration of DNA when recording the results after PCR and gel electrophoresis. A smear of DNA is observed in the well where the amplified DNA is loaded, instead of a discrete DNA band.
- Use of in-house PCR methodologies. Some diagnostic laboratories do not use any of the OIE-recommended procedures. Instead, they use their own PCR protocol developed in-house. Sometimes these procedures are not optimized and validated properly, resulting in failure to amplify the target pathogens.
False positive
The issues related to false positives occur due to:
- Cross contamination during the sampling process. Due to a high viral/bacterial load in a sample, cross contamination can happen.
- Nested PCR. Some laboratories use a nested PCR for some pathogen detection. During the nested PCR step, a positive sample with a high pathogen load can contaminate subsequent negative samples due to pipetting error and/or formation of aerosols during PCR set-up. These errors will cause a cross contamination.
- A non-specific band with a similar size to a specific band. Due to a lack of proper optimization, some PCR protocols result in non-specific amplification with a DNA band similar to the size of a positive control. Some laboratories might consider these non-specific bands as positive.
Perspectives
Diagnosis of shrimp pathogens is a critical component of the shrimp industry. Therefore, a reliable diagnostic laboratory must be an integral part of the biosecurity strategy to ensure the sustainability of the shrimp industry worldwide. The data we have collected over the years clearly show that participation in the PT conducted by the University of Arizona Aquaculture Pathology Laboratory can facilitate in improving the performance of a shrimp diagnostic laboratory worldwide.
Authors
-

Fernando Aranguren Caro, Ph.D.
Aquaculture Pathology Laboratory
School of Animal and Comparative Biomedical Sciences
University of Arizona
1117 E Lowell St.
Tucson, Arizona 85721 USA -

Jasmine Millabas
Aquaculture Pathology Laboratory
School of Animal and Comparative Biomedical Sciences
University of Arizona
1117 E Lowell St.
Tucson, Arizona 85721 USA -

Arun K. Dhar, Ph.D.
Aquaculture Pathology Laboratory
School of Animal and Comparative Biomedical Sciences
University of Arizona
1117 E Lowell St.
Tucson, Arizona 85721 USA
Tagged With
Related Posts

Health & Welfare
Big shoes to fill: Dhar takes reins at shrimp pathology laboratory
Arun Dhar, Ph.D. will attempt to fill the “big shoes” of Dr. Donald Lightner at the University of Arizona’s Aquaculture Pathology Laboratory, where the shrimp disease EMS was diagnosed.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
A holistic management approach to EMS
Early Mortality Syndrome has devastated farmed shrimp in Asia and Latin America. With better understanding of the pathogen and the development and improvement of novel strategies, shrimp farmers are now able to better manage the disease.

Health & Welfare
Four AHPND strains identified on Latin American shrimp farms
Two virulence genes are known to encode a binary photorhabdus insect-related toxin that causes acute hepatopancreatic necrosis disease in shrimp. The pathogenicities of these V. campbellii strains were evaluated through laboratory infection and subsequent histological examination in P. vannamei shrimp.

