Barramundi case study demonstrates potential of novel tool

Diseases are a major impact on, and represent one of the major constraints to future growth of, the aquaculture sector. It has been reported that diseases are responsible for approximately 40 percent of production losses in aquaculture ($102 billion). Therefore, innovative approaches are needed for the early detection and management of disease outbreaks in aquaculture farms. Environmental DNA (eDNA) is a sampling approach associated with molecular techniques that allows the detection of the genetic material of any micro- or macro-organism present in the environment. This technique has the potential to revolutionize the way pathogens are detected and monitored in aquaculture.
Environmental DNA presents an alternative to invasive sampling and quantification of pathogens by collecting water or soil samples from aquaculture systems. While traditional diagnostics (e.g. histopathology, microbiology, PCR) continue to be the gold standard methodologies to confirm the causative agent of infectious diseases in animals, these techniques are time consuming. By the time farmers obtain results from traditional diagnostic tests to control outbreaks. The environmental DNA approach offers the opportunity to improve animal health monitoring by detecting pathogens in aquaculture systems before animals get infected.
Among different pathogens affecting the aquaculture industry, ciliate protozoans are considered some of the most economically important parasites of finfish. Generally, ciliate parasite outbreaks progress rapidly, without any warning, causing mortalities very quickly. Chilodonella hexasticha is a common ciliate parasite found in freshwater finfish systems, which can alternate between parasitic and free-living stages, killing fish within 2-3 days. Using C. hexasticha as model organism, the eDNA methodology was tested to early detect and quantify pathogens in a barramundi or Asian seabass (Lates calcarifer) farm.
Note: This article is adapted and summarized from the original publication (Aquaculture 479, 2017).
Barramundi case study: sampling and laboratory processing
Water samples from eight earthen ponds (~1.4 ha) of a commercial freshwater barramundi farm from Australia were collected monthly over one year. Triplicate 15 mL pond water samples were collected using individual plastic cups and poured into 50 mL centrifuge tubes containing 1.5 mL of sodium acetate (3 M) and 33.5 mL of absolute ethanol for DNA preservation. Samples were stored on ice until processing in the laboratory.
Samples were processed using a direct eDNA precipitation and extraction method where the genetic material was extracted using a CTAB extraction protocol. Ciliate parasite cells from infected barramundi were collected during the same period for species identification and validation of a quantitative PCR (qPCR) assay. The qPCR assay was developed to identify C. hexasticha abundance (copies cell/µl) in water samples based on the species SSU-rDNA gene region. Figure 1 shows a sample collection flow diagram, from water collection on farm to processing in the molecular laboratory.
Additionally, environmental and biological data monitored on the farm (dissolved oxygen, temperature, rainfall, fish weight and mortalities) were associated with the abundance of C. hexasticha cell copies/µl (eDNA) in water. This relationship between water quality parameters and parasite abundance was tested using the mean maximum water temperature, minimum dissolved oxygen levels and rainfall over a consecutive 5-day period prior to water sampling for each pond. Similarly, the relationship between parasite abundance in water and fish mortality was tested using the mean number of dead fish recorded over the 5-day period following water sampling in each pond.
Analyses were performed through linear regression and correlation to predict fish mortalities observed in the 5 days post water sampling using measured variables (i.e. abundance of C. hexasticha (eDNA), rainfall, dissolved oxygen and water temperature and fish weight) and to evaluate whether any of the observed environmental variables could predict the abundance of C. hexasticha in water samples. Additionally, a principal component analysis (PCA) was used to determine the association between the observed variables and the abundance of parasite cells in pond water.
Results and discussion
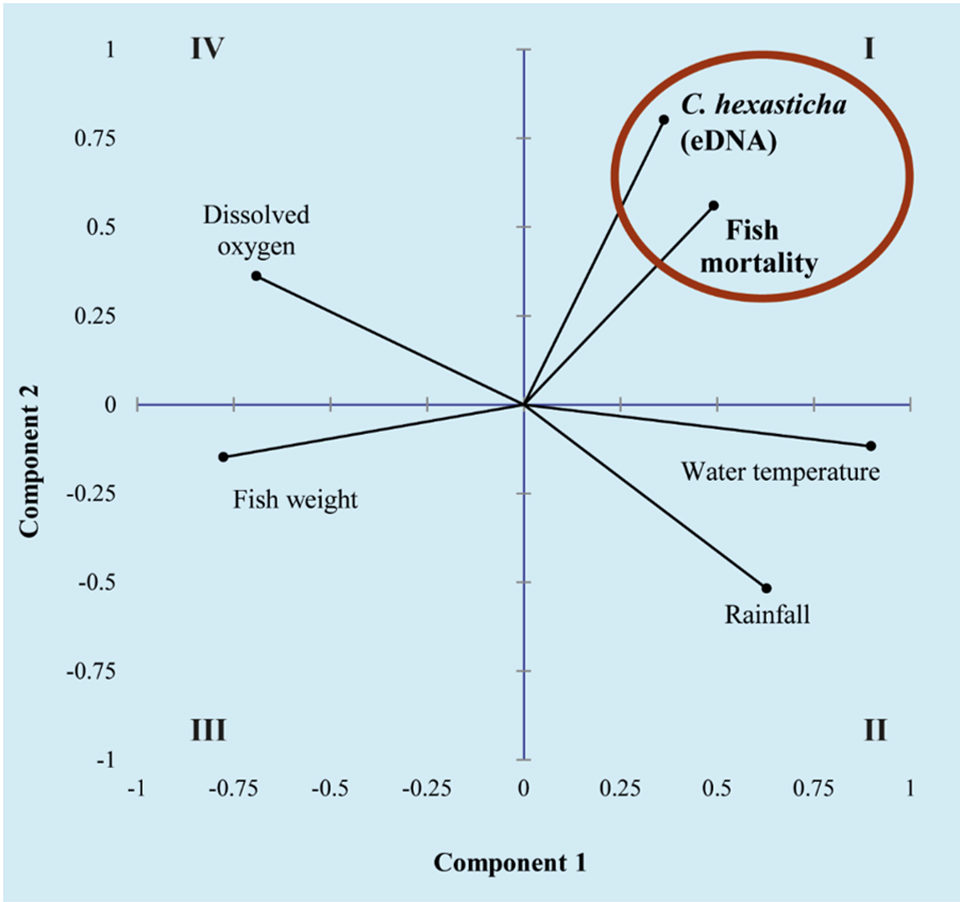
This study used an eDNA approach to quantify the abundance of the ciliate parasite C. hexasticha (eDNA) in water from a commercial barramundi farm and investigate its relationship with water quality and production data. A positive association was observed between abundance of C. hexasticha in pond water (eDNA) and fish mortalities (red circle; quadrant I; Fig. 2), which were inversely related to fish weight (quadrant III; Fig. 2) and dissolved oxygen levels (quadrant IV; Fig. 2). This was further confirmed by a positive correlation between abundance of parasite in pond water (eDNA) and fish mortalities but negatively correlated with fish size. Fish mortalities were also strongly and positively correlated with warmer water. In other words, in the studied farm smaller barramundi tended to be particularly affected by high levels of the ciliate parasite in water and mortalities occurred during the rainy and warm season.
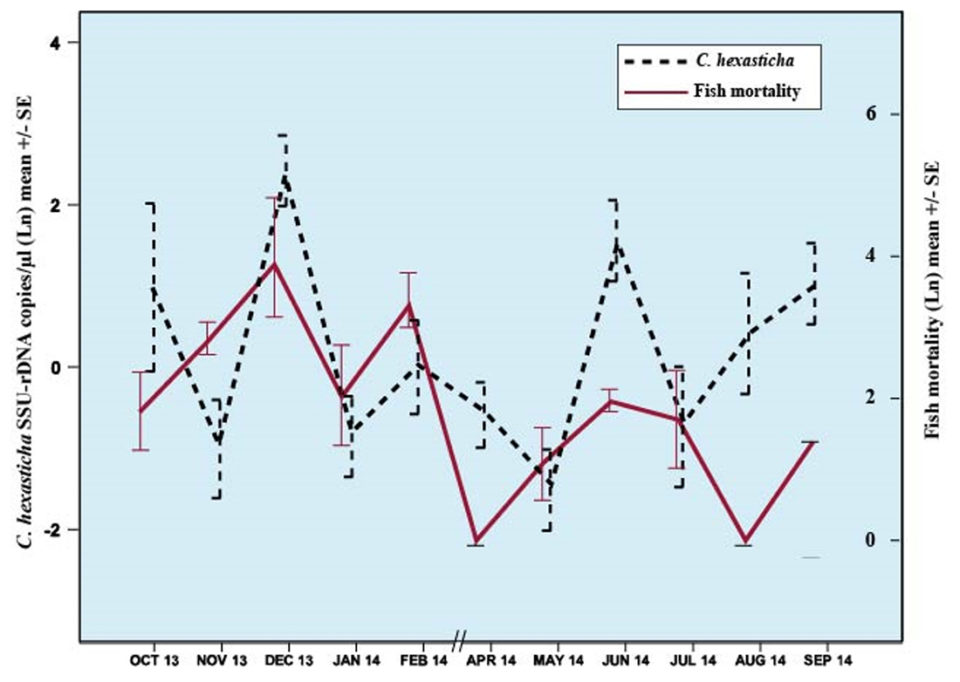
Interestingly, a synchronicity between abundance of C. hexasticha cells in water (eDNA) and observed fish mortalities five days post sampling was observed (Fig. 3). Samples from months with higher abundance of parasite in water (eDNA) were also months with higher fish mortalities (Fig. 3). The high abundance of C. hexasticha cells in water and fish weights were significant predictors of fish mortalities, while lower rainfall and fish weights were significantly associated with the high abundance of C. hexasticha cells in water.
Environmental DNA showed to be a powerful approach that can be used to predict fish mortalities, especially when associated with environmental parameters recorded on farm. Given the fact that usually severe fish mortalities occur in fish farms without warning, the quantification of parasite DNA from water samples provide production managers and health professionals with a new efficient tool to assess the risk of future disease outbreaks.
While the eDNA approach for pathogen monitoring in aquaculture is a new area of research, its potential is still under-explored in commercial conditions. Environmental DNA approaches can be used as a non-invasive technique for pathogen sampling to minimize stress and improve animal wellbeing in aquaculture enterprises.
Considering that limited knowledge exists on the diversity of microbial communities within aquaculture systems, eDNA represents a new platform for the industry to unleash the secrets under the water. More specifically within the aquaculture animal health sector, eDNA offers the possibility to understand the abundance and co-infections of pathogens in culture systems. Using next-generation and high throughput sequencing techniques, one single assay can be developed to identify the presence and abundance of multiple aquatic communities in farms and hatcheries.
Finally, eDNA can be easily performed on-farm with the use of portable devices and be incorporated into routine monitoring programs as an efficient tool to assess the risk of future disease outbreaks, thus allowing fish farmers to promptly adopt preventative health management and early intervention strategies.
Authors
-
Giana Bastos Gomes, Ph.D.
Corresponding author
James Cook University Singapore
Singapore, 387380 and
Marine Biology and Aquaculture Sciences
College of Science and Engineering and
Centre for Sustainable Tropical Fisheries and Aquaculture
James Cook University
Townsville, 4811, QLD, Australia[117,97,46,117,100,101,46,117,99,106,64,115,101,109,111,103,46,97,110,97,105,103]
-
Kate S. Hutson, Ph.D.
Marine Biology and Aquaculture Sciences
College of Science and Engineering and
Centre for Sustainable Tropical Fisheries and Aquaculture
James Cook University, Townsville, 4811, QLD, Australia -
Jose S. Domingos, Ph.D.
James Cook University Singapore
Singapore, 387380, and
Marine Biology and Aquaculture Sciences
College of Science and Engineering and
Centre for Sustainable Tropical Fisheries and Aquaculture
James Cook University, Townsville, 4811, QLD, Australia -
Catherine Chung
Sealord King Reef
188 Cowley Creek Road
Cowley, 4871, QLD, Australia -
Scott Hayward
Sealord King Reef
188 Cowley Creek Road
Cowley, 4871, QLD, Australia -
Terrence L. Miller, Ph.D.
Fish Health Laboratory
Department of Fisheries Western Australia
South Perth 6151, WA, Australia -
Prof. Dean R. Jerry, Ph.D.
James Cook University Singapore
Singapore, 387380, and
Marine Biology and Aquaculture Sciences,
College of Science and Engineering and
Centre for Sustainable Tropical Fisheries and Aquaculture
James Cook University, Townsville, 4811, QLD, Australia
Tagged With
Related Posts

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Health & Welfare
A comprehensive look at the Proficiency Test for farmed shrimp
The University of Arizona Aquaculture Pathology Laboratory has carried out the Proficiency Test (PT) since 2005, with 300-plus diagnostic laboratories participating while improving their capabilities in the diagnosis of several shrimp pathogens.

Intelligence
Adding flavor complexity to farmed barramundi
Organoleptic attributes such as flavor and aroma are among the most important factors that influence consumer acceptability and demand for fish products. Consumers have identified farmed fish as less complex and lacking “sealike” or “sea-fresh” flavors and aromas.

Health & Welfare
Calling for a National Broodstock Improvement Network
A National Broodstock Improvement Network for aquaculture could help to break the link between inbreeding and hatchery size.


