Variations in salinity tolerance can be used to develop hybrids

With the increasing scarcity of freshwater available for aquaculture in general, and for tilapia culture in arid regions like Israel in particular, the development of fish that tolerate high salinity would increase global tilapia output by expanding the range of production in many regions of the world.
The authors recently reviewed the subject, aiming to show what can be learned from past experiences in culture management practices and nutrition, as well as physiology and genetics.
Salt tolerance
The control of salt and water balance within a narrow limit is critical to life in all multicellular organisms, including teleost fish. Salt tolerance is a term that describes the overall fitness or productivity of fish in a saline environment. It is a combination of different quantitative traits, such as metabolism, growth, osmoregulation, immunocompetence and fecundity. Existing interspecific variations in salinity tolerance can be used to select salt-tolerant species and develop salt-tolerant hybrids.
Chloride cells
Chloride cells, also known as mitochondrion-rich cells, in the gill epithelium are important osmoregulatory sites in all fish species. Their large surface area at both sides, the apical and basolateral, are locations for ion-transporting proteins such as sodium-potassium ATPase and sodium-potassium chloride co-transporter.
Studies, mostly in Oreochromis mossambicus, on changes in chloride cells’ characteristics and functions in response to salinity challenges revealed a significant increase in the abundance of chloride cells and ion transporter activity in the gills. Differences in ion transporter type and membrane location on the chloride cells were also found between freshwater- and saltwater-challenged fish.
Hormones, osmoregulation
Hormones of the neuroendocrine system are essential players in the control of osmoregulatory mechanisms, and extensive studies on endocrine pathways involved in osmoregulation clarified a significant role of prolactin (PRL) and growth hormone (G.H.) in osmoregulation.
Growth and development are directed by an integration of environmental, physiological and genetic factors. The high energetic cost of osmoregulation, usually estimated at 25 to 50 percent of metabolic output, means there is a link between osmoregulatory and growth capacities. This might be related to the observation that growth and osmoregulation are governed by many of the same hormones, notably PRL and G.H. It has been demonstrated that genetic variation in the tilapia PRL gene is associated with differential gene expression and growth rate in saline water.
Genomics tools
Functional genomics – the field of molecular biology that attempts to answer questions about the function of DNA at the levels of genes, RNA transcripts and protein products – and proteomic approaches that study the structures and functions of proteins represent powerful tools for gaining insight into the molecular bases of environmental adaptation.
Gene transcripts for ion transporters, enzymes, hormones and components of cellular stress signaling were characterized in the brains, gills, guts and kidneys of Mozambique tilapia (O. mossambicus) and black-chinned tilapia (Sarotherodon melanotheron). Most genes showed an immediate response with peak levels observed two to eight hours after seawater transfer.
Pathway analysis of the newly identified genes revealed that more than half of the immediate hyperosmotic stress genes interacted closely within a cellular stress response signaling network.

Approaches to salt tolerance
Salt tolerance in tilapia can be improved by various approaches, such as optimizing acclimation protocols and adding salt to the diet. Exploiting existing variations within and among species, salt-tolerant strains can be produced through hybridization between fast-growing but less salt-tolerant species, such as Nile tilapia, and salt-tolerant ones, such as Mozambique tilapia.
Salt tolerance can also be improved by selective breeding. The results from a few pioneer projects suggested that substantial additive genetic variance exists for growth rate and survival in saline environments that can be exploited through selective-breeding programs.
Finally, the application of genomic approaches and modern molecular biology techniques may enable the identification of genes that encode specific proteins active in salt-tolerant species that are lacking or less active in less-tolerant species, or specific proteins that are induced under salt stress.
Prolactin 1 gene
One such gene is prolactin 1. It has a central role in the adaptation of marine species to freshwater by reducing sodium-potassium ATPase activity and consequently increasing the osmotic level of the plasma. Microsatellite polymorphism in the tilapia prolactin 1 promoter was shown to be associated with differences in prolactin 1 gene expression and growth response in salt-challenged fish.
Fish homozygous for the long allele grew slower at 16 ppt, and their weight was only half those of the heterozygotes and homozygotes for the short allele, while in freshwater, growth rate did not differ significantly among the three genotypes.
Genetics, environment
The authors recently re-examined this association in nine F2 families of O. mossambicus x O. niloticus hybrids. Both parental fish were heterozygous for different alleles (CA33 and CA38 in O. mossambicus, CA30 and CA35 in O. niloticus, resulting with polymerase chain reaction products of 253, 263, 247 and 257 bp, respectively). The authors confirmed this association in three out of nine F2 families of O. mossambicus x O. niloticus hybrids in saline water. The six non-segregating families were from a different spawning season.
The same pattern of improved growth for genotypes with shorter alleles originating from the O. niloticus grandparental fish was demonstrated, although O. mossambicus is considered a more salt-tolerant species. In two of the three families, full-sibs were also grown in freshwater, where no correlation between the genetic polymorphism and growth was found.
In these two families, fish carrying the allelic combination 247/253 grew better in saline water and worst in freshwater (Figure 1). The effects accounted for only 13 to 15 percent of the phenotypic variance for growth rate (P < 0.05).
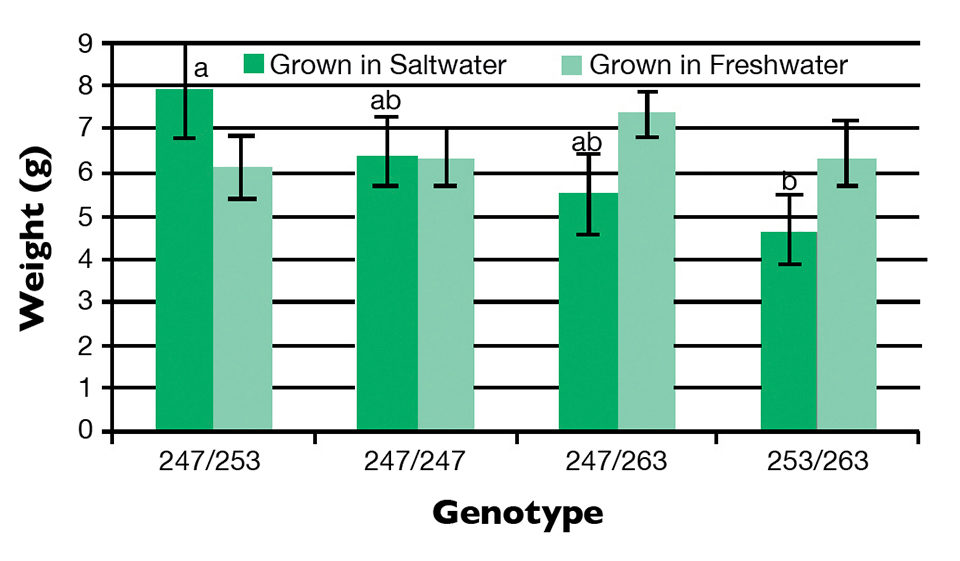
The authors concluded that this variation is probably not a major contributor to the total genetic variation in salinity tolerance, and there may also be a large environmental influence underlying the differential growth in saline water. Nevertheless, no association was evident between the polymorphism in the prolactin 1 promoter and the expression of the gene.
Perspectives
Several genes that are differentially expressed in tilapia in saltwater and freshwater – beta haemoglobin, ATPase Ca2þ+ -transporting plasma membrane ATPase, pro-opiomelanocortin (all upregulated in saltwater) and beta-actin (downregulated) – were identified as candidate genes associated with salt tolerance.
Transferrin, an iron-binding glycoprotein known to have an important role in the immune system, also showed an 85 percent upregulation in tilapia kept in saltwater compared to freshwater, suggesting it may be involved in saltwater tolerance or that closely linked genes may be directly involved in saltwater tolerance.
In conclusion, studies on the molecular basis of osmoregulatory properties of the gills, kidney, gut and brain have revealed a wealth of genomic knowledge that can lead to genetic studies of intra- and interspecific variation for salinity tolerance. Once relevant genes are identified, genetic polymorphisms can be sought in cultured and natural populations.
The emerging knowledge of quantitative trait loci associated with, or genes directly involved in saltwater tolerance may facili-tate marker-assisted or gene-assisted selection for this trait in tilapia in the future. This will become even more efficient, now that the first draft of the tilapia genome has been released.
Two routes hold the keys for improving salinity tolerance. First, exploring and revealing biochemical pathways and gene networks involved in osmoregulation can realize better understanding of both the salt tolerance phenotype and the genotypic background. Screening domesticated and natural populations, searching for genetic variations in the biochemical pathways that underlie the observed phenotypic differences, is another route.
Knowledge so gained can be exploited in the selective breeding of tilapia stocks that perform well in saline waters.
(Editor’s Note: This article was originally published in the November/December 2011 print edition of the Global Aquaculture Advocate.)
Authors
-
Avner Cnaani, Ph.D.
Institute of Animal Science
Agricultural Research Organization
Volcani Center
P. O. Box 6
Bet Dagan 50250, Israel[108,105,46,118,111,103,46,105,114,103,97,46,105,110,97,99,108,111,118,64,99,114,101,110,118,97]
-
Ariel Velan, M.S.
Institute of Animal Science
Agricultural Research Organization
Volcani Center
P. O. Box 6
Bet Dagan 50250, Israel -
Gideon Hulata, Ph.D.
Institute of Animal Science
Agricultural Research Organization
Volcani Center
P. O. Box 6
Bet Dagan 50250, Israel
Tagged With
Related Posts

Aquafeeds
A look at phospholipids in aquafeeds
Phospholipids are the major constituents of cell membranes and are vital to the normal function of every cell and organ. The inclusion of phospholipids in aquafeeds ensures increased growth, better survival and stress resistance, and prevention of skeletal deformities of larval and juvenile stages of fish and shellfish species.

Innovation & Investment
A review of unit processes in RAS systems
Since un-ionized ammonia-nitrogen and nitrite-nitrogen are toxic to most finfish, controlling their concentrations in culture tanks is a primary objective in the design of recirculating aquaculture systems.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 2
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors considered biotic and abiotic factors. Part 2 describes results of their study.

Health & Welfare
Acclimating shrimp postlarvae before pond stocking
Shrimp postlarvae acclimation before stocking into the various growout systems (ponds, raceways, tanks) is a critical – and often overlooked, sometimes taken for granted – step in the shrimp culture process. Various water quality parameters should be changed slowly so that the young shrimp have the time to gradually adapt to the new conditions.


